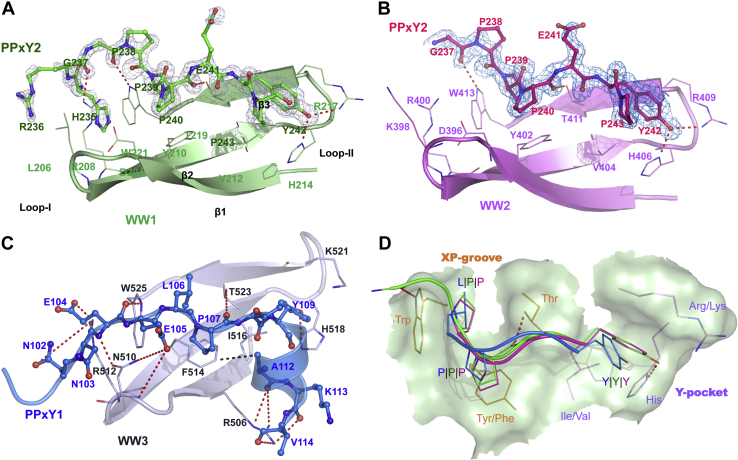
Figure 4.
Structures of AMOT PPxY–NEDD4L WW complexes. A, crystal structure of AMOT PPxY2 (green sticks, chain b) bound to NEDD4L WW1 (pale green, chain a; ribbon diagram with key residue side chains shown explicitly). The three WW domain strands are labeled in this panel. B, crystal structure of AMOT PPxY2 (magenta, chain d) bound to NEDD4L WW2 (pink, chain c). Red dashed lines in panels A and B denote observed hydrogen bonds. Mesh renderings show the FO-FC map (3σ), calculated with the PPxY peptide omitted. C, solution structure of AMOT PPxY3 (blue) bound to NEDD4L WW3 (gray). Dashed red lines indicate observed hydrogen bonds or salt bridge interactions. The hydrophobic pocket interaction between WW3 Phe514 and PPxY1 Ala112 (∼3.9 Å) is shown as black dashed lines. Glu105 interacts with Asn510 (2.7–4.5 Å) in ∼50% of ensemble structures. Each of the residues Glu104 and Asn102 individually interacts with Arg512 (2.6–4.5 Å) in ∼40% of ensemble structures. Carbonyl oxygen of Glu105 hydrogen bonds to side-chain nitrogen of Asn510 (not shown for clarity). Arg506 interacts with the carbonyls of Ala112, Lys113, and Val114 (3.0–4.5 Å) in ∼25% of ensemble structures. D, surface rendering of the conserved WW core domain showing the characteristic XP groove (residue labels in orange) and Y-pocket (residue labels in purple). PPxY peptides are overlaid and colored following panels A-C. Red dashed lines denote canonical WW–PPxY hydrogen bonds (shown only for WW1–PPxY2 for clarity). PPxY, Pro-Pro-x (any amino acid)-Tyr.