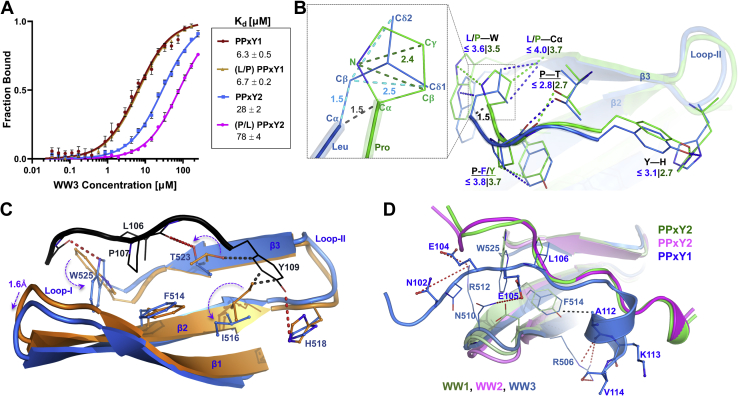
Figure 5.
Energetic and structural consequences of NEDD4L WW3 binding to wt and mutant AMOT PPxY motifs.A, fluorescence polarization (FP) binding isotherms and fitted Kd values (μM) for NEDD4L WW3 binding to peptides corresponding to wt AMOT PPxY1 (red), an PPxY1 L106P mutant (orange, denoted (L/P)PPxY1), wt PPxY2 (blue), and a PPxY2 P239L mutant (magenta, denoted (P/L)PPxY2). Error bars denote SD from four independent replicates. B, superpositions of PPxY1–WW3 (blue) and PPxY2–WW1 (green) complexes illustrating how the noncanonical Leu106 residue of the PPxY1 occupies the same position as the canonical Pro239 of PPxY2. Key conserved residues are shown explicitly; AMOT PPxY–WW distances (dashed lines, Å) are shown. WW3–PPxY1 distances are from ∼75% of ensemble structures. Inset: close-up view of Leu/Pro residues within the boxed region. Note that the Pro pyrrolidine ring and the Leu isopropyl group occupy the same WW binding site (Trp, Thr, Tyr/Phe, and His) and bury similar accessible surface areas (∼90 Å2). Distances (Å, dashed lines) are shown for scale and to illustrate the 1.5 Å shift between the PPxY1 Leu106 and PPxY2 Pro239 Cα positions. Substitution of Leu for Pro in the first position of the PPxY motif should reduce the propensity to form a PPII helix (37), but this unfavorable change may be compensated by burial of the larger Leu side chain. C, superposition of WW3 structures in the apo/unliganded (orange) and PPxY1 (blue) states. Displacements of the loop 1 and key side chains upon peptide (black) binding are shown as purple arrows and dashed lines. Red dashed lines depict canonical hydrogen bonds, and black dashed lines show steric clashes that require WW3 Ile516 (1.5 Å to Tyr109) and Thr523 (3.0 Å to Tyr109) to change conformation. D, superposition of PPxY2–WW1 (green), PPxY2–WW2 (magenta), and PPxY1–WW3 (blue), emphasizing the interactions unique to WW3–PPxY1. PPII, polyproline II; PPxY, Pro-Pro-x (any amino acid)-Tyr.