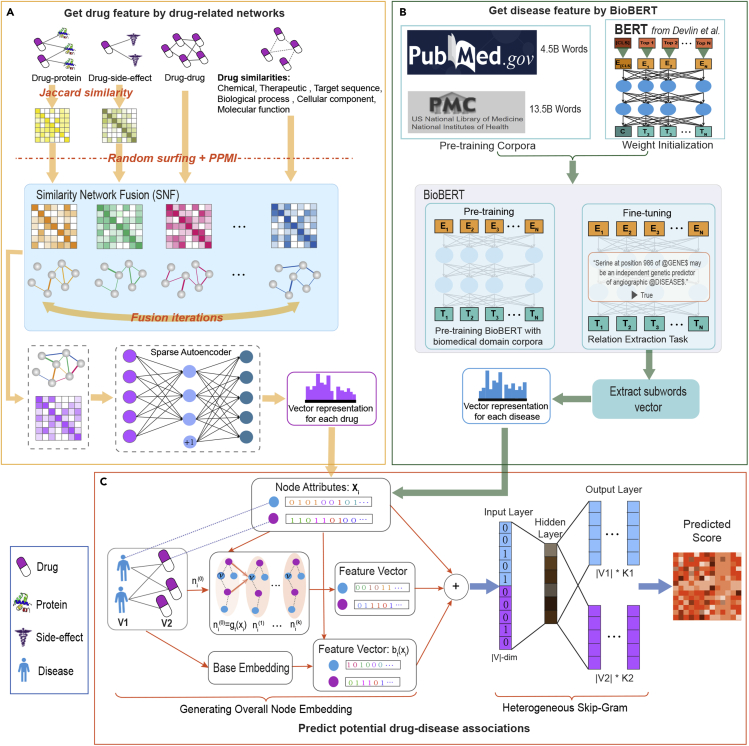
Figure 1.
Flowchart of HeTDR
The model consists of three parts: (A) HeTDR integrates nine drug-related networks to obtain global information of drugs. In the heterogeneous interaction networks, we first use the Jaccard similarity coefficient to calculate the similarity network. Then, we fuse these drug-related networks into one network by SNF and apply SAE to obtain low-dimensional features of the drugs. (B) HeTDR obtains vector representation of the disease features by text mining biomedical corpora. In the pre-training stage, we directly use the model parameters pre-trained by BioBERT. Then, we select the relation extraction task for fine-tuning training. After the fine-tuning process has taken place, we extract the representations of sub-words and use the representations of these sub-words to obtain the representations of all diseases. (C) HeTDR predicts potential drug-disease associations by an embedding learning method, which can capture both the drug-disease associations network topological structural proximity and node attributes proximity.