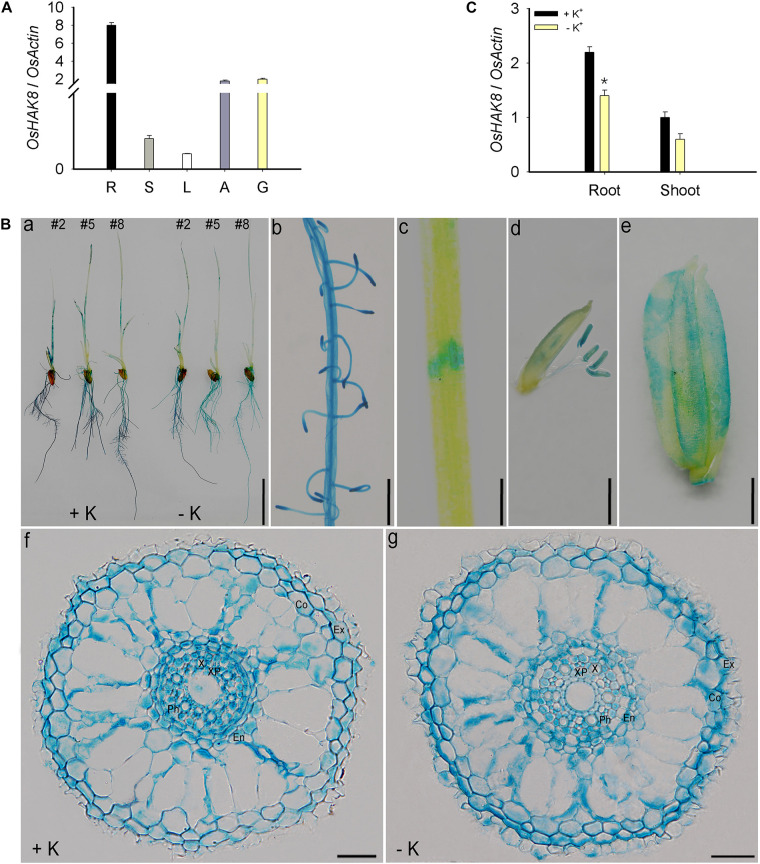
FIGURE 5.
Expression pattern of OsHAK8. qRT-PCR analyses showing OsHAK8 mRNA accumulation in roots, stems, leaves, anthers, and glumes. Rice seedlings (Nipponbare) were grown in soil for 10 weeks. R, root; S, stem; L, leaf; A, anther; G, glume. (A) Histochemical analysis of OsHAK8 promoter-driven GUS reporter expression in transgenic rice plants. The transgenic plants were grown as described in (B), and GUS activities (indicated in blue) were examined using histochemical staining. (a) GUS staining of 7-days-old plants (Line 2) grown in hydroponic solutions with 10-mM K+ (+ K) or 0-mM K+ (-K). (b) Enlarged image of the root hair zones in (a). (c) Enlarged image of the node in (a). (d) GUS staining of anthers. The transgenic plants were grown as described in (A). (e) GUS staining of glume. The transgenic plants were grown as described in (A). (f,g) Cross-section images of the elongation zone in (a). Ex, exodermis; Co, cortex; En, endodermis; Ph, phloem; X, xylem; XP, xylem parenchyma. Bar in (a) = 1 cm, bar in (b) = 5 mm, bar in (c) = 1 mm, bar in (d) = 1 mm, bar in (e) = 1 mm, and bars in (f) to (g) = 100 μm. (C) Effect of K+ deficiency on the transcriptional expression of OsHAK8 in rice. 3-day-old rice seedlings (Nipponbare) were grown in a hydroponic solution containing 10 mM K+ for 3 days and then transferred to 10-mM K+ (+ K) and 0-mM K+ (−K) solutions for 24 h, respectively. Total RNAs were isolated from the roots and shoot of the seedlings, and OsHAK8 transcript levels were determined by qRT-PCR. Actin was used as an internal standard. Significant difference was found between 0- and 10-mM K+ samples indicated in root tissue (P<0.01 by Student’s t-test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Asterisks represent significant differences.