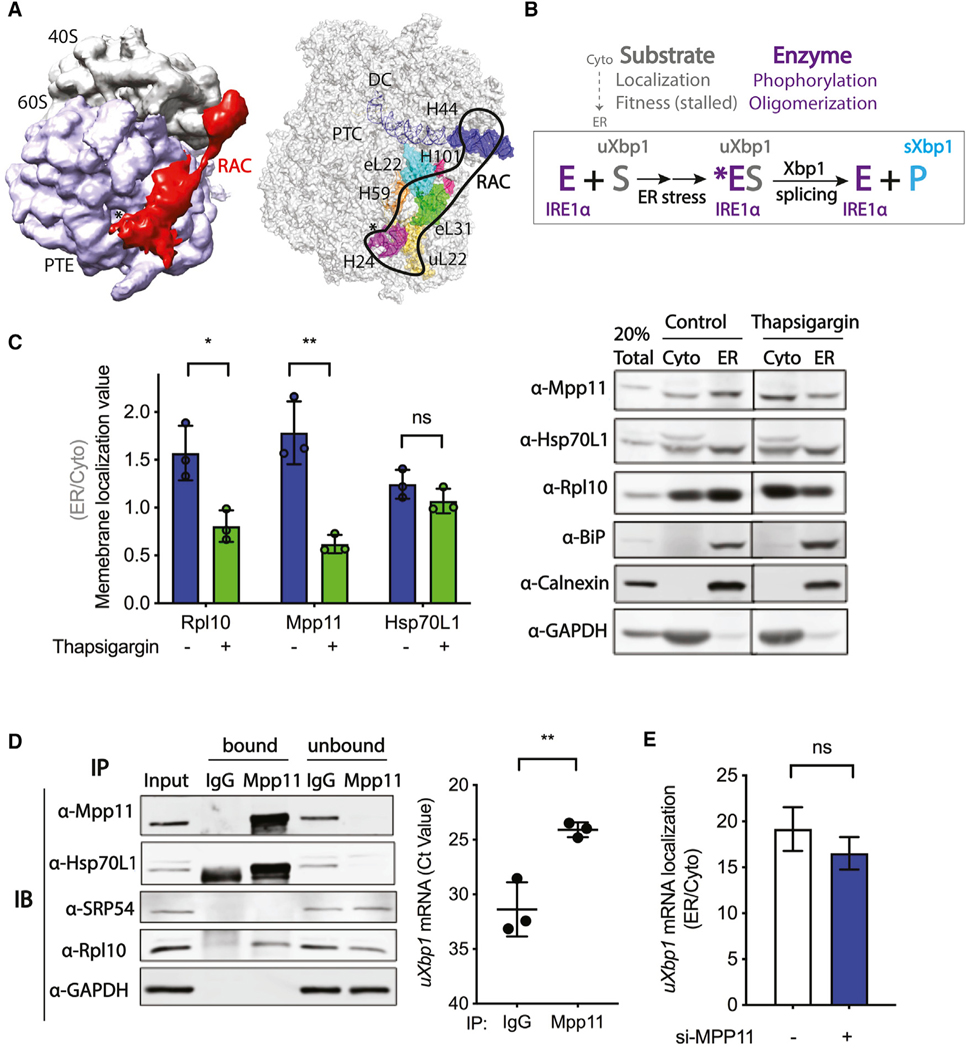
Figure 2. RAC is not required for membrane targeting of the uXbp1 mRNA-ribosome substrate of the IRE1α-catalyzed splicing reaction.
Probable mechanisms of RAC regulation on Xbp1 mRNA splicing.
(A) (left) Cryo-EM density map of RAC with the 80S ribosome in S. cerevisiae by Chimera (EMD: 6103). RAC contacts both the 40S and 60S ribosomal subunits. RAC, red; 40S, gray; 60S, lavender; *, PTE, polypeptide tunnel exit. (right) Interaction sites of RAC with the 80S ribosome by PyMOL (PDB: 3J78). On the 60S subunits, RAC binds near the PTE by ribosomal protein eL22, uL22, and H24; H59 of 28S rRNA; as well as eL31 and H101. On the 40S subunits, RAC contacts the ES12 of H44 of 18S rRNA, which stems from the decoding center (DC) (Leidig et al., 2013; Yan et al., 1998; Zhang et al., 2014). RAC, black outline; *, PTE; eL31, green; H101, hot pink; H24, purple; H59, orange; eL22, cyan; uL22, yellow; H44, blue; PTC, the peptidyl transferase center.
(B) The Xbp1 splicing reaction. Under basal conditions, the IRE1α kinase-endonuclease E is in an inactive monomeric state on the ER. The substrate uXbp1 mRNA S requires several steps to be fit for the IRE1α enzyme and formation of the ES complex. First, translation must be initiated and then stalled at the proper location and the ribosome-uXbp1 is transported by SRP to the translocon on the ER membrane near where the IRE1α enzyme is located. The enzyme must also be activated. Upon ER stress (*), IRE1α is phosphorylated and then oligomerizes in foci. The active enzyme acts on the fit substrate to splice out the intron of uXbp1, releasing the product sXbp1 P. RAC could modulate Xbp1 splicing by affecting any of these steps.
(C) Subcellular fractionation of HeLa cells harvested after 4 h of DMSO control (blue bar) or thapsigargin (0.5 μM) (green bar) treatment was carried out using the sequential detergent extraction method. A total of 20% of total lysate was loaded; cytosol (Cyto)- and ER-membrane-bound fractions were collected and loaded in equal amounts. Quantification of western blots (left) for RAC and other components of the system used to determine a membrane localization value (ER/Cyto). Representative western blot images of subcellular fractionation (right). Rpl10 was used as a marker for ribosomes, BiP as a marker for ER lumen, Calnexin as marker for ER membrane, and GAPDH as a marker for cytosol. n = 3; *p < 0.05, **p < 0.01. Error bars, mean ± SD.
(D) RNA immunoprecipitation was performed in HeLa cells. Lysates were immunoprecipitated with antibody against IgG control or Mpp11, and the co-precipitated RNA was determined by qRT-PCR. (left) Representative western blot of bound and unbound proteins. A total of 1% total lysate was used as input. (right) uXbp1 mRNA is shown as Ct value. n = 3; **p < 0.01. Error bars, mean ± SD.
(E) Subcellular fractionation of RNA by the differential detergent method in HeLa cells pretreated with either vehicle control (white) or Mpp11 (blue) siRNA for 48 h.RNA was analyzed by qRT-PCR; samples were compared to vehicle si-control and calculated as 2−ΔCT. uXbp1 mRNA localization is shown as ER/cytosol. n = 2. Error bars, mean ± SEM.