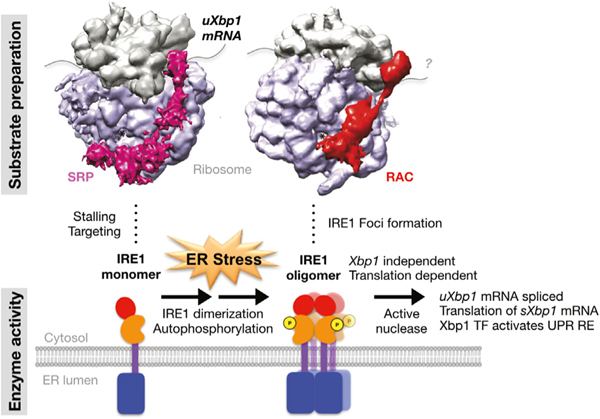
Figure 7. A model for RAC’s role in IRE1α oligomerization and activation.
Schematic of the steps at which RAC influences the IRE1α branch of UPR. Under basal conditions, the monomeric IRE1α kinase-endonuclease is stabilized in an inactive form by binding to BiP. Translation of the IRE1α’s substrate uXbp1 mRNA is initiated and stalled on cytosolic ribosomes, presumably by SRP (magenta), which targets the substrate complex from the cytosol to the ER membrane. Upon ER stress, unfolded proteins compete with IRE1α for BiP and release of BiP allows IRE1α to dimerize, activating its kinase activity and mediating production of the autophosphorylation, which can catalyze RIDD. The dimerized IRE1α can further oligomerize, forming foci that correlate with the activation of the splicing endonuclease activity catalyzed by IRE1α. The cryptic exon is spliced from uXbp1, stalling relieved, translation terminated, and the sXbp1 released from the ribosome. The translation of sXbp1 mRNA is reinitiated, producing the Xbp1 transcription factor, which then activates expression of UPR genes. RAC (red) plays an unexpected role in activation of UPR by modulating the IRE1α high-order oligomerization required for activity. Activation depends upon translation but independently of Xbp1. The similarities in the RAC ribosome and SRP ribosome complex and their structural relationship to the nascent chain exit tunnel and the decoding center are shown in cryo-EM density maps of SRP (EMD: 1063) or RAC (EMD: 6103) with the 80S ribosome produced with Chimera. The IRE1α enzyme activation and the substrate preparation processes are indicated below the structure by the cartoons. IRE1α domains: red, RNase; orange, kinase; purple, transmembrane; blue, luminal domain.