Figure 19.
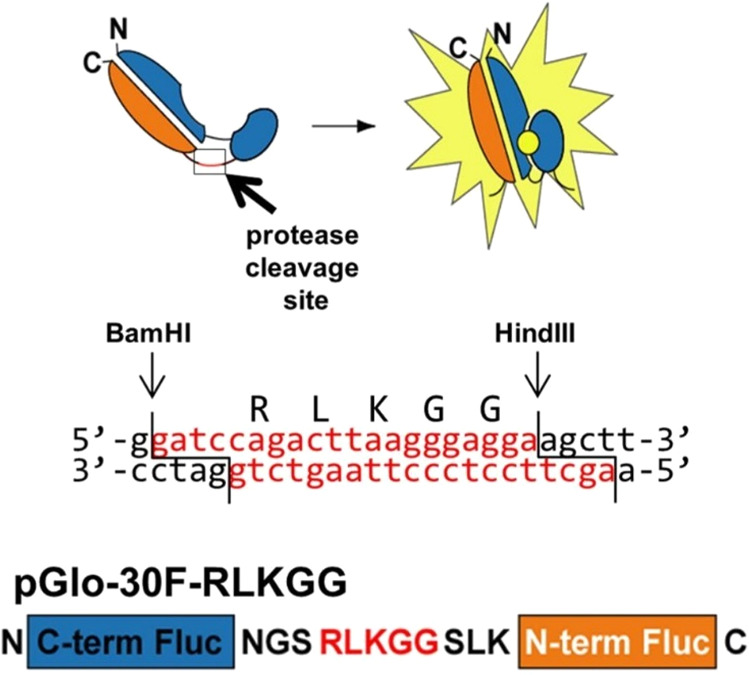
Schematic representation of the circularly permuted luciferase construct linked by the amino acid sequence RLKGG for assessing MERS-CoV papain-like protease (PLpro) activity in cells. For coronaviruses including MERS-CoV, PLpro processing of viral polyproteins is responsible for the formation of the MERS-CoV replicase complex following host cell entry and viral mRNA translation. Effective inhibitors of PLpro will lead to inhibition of MERS-CoV replication and will thus show little to no bioluminescence in the presence of the circularly permuted luciferase construct and the protease. This assay is effective for evaluating protease activity and to screen for potential inhibitors. pGlo-30F: vector backbone. RLKGG: coronavirus (CoV) papain-like protease (PLpro) cleavage site. BamHI, HindIII: restriction enzyme cleavage sites. While sharing high sequence identity, the PLpro of pathogenic coronaviruses such as SARS-CoV and SARS-CoV-2 exhibit distinct host substrate preferences.96 Nonetheless, it may be possible to identify a PLpro inhibitor that can offer broad suppression of coronavirus replication. Adapted with permission from ref (95). Copyright 2013 American Society for Microbiology.
