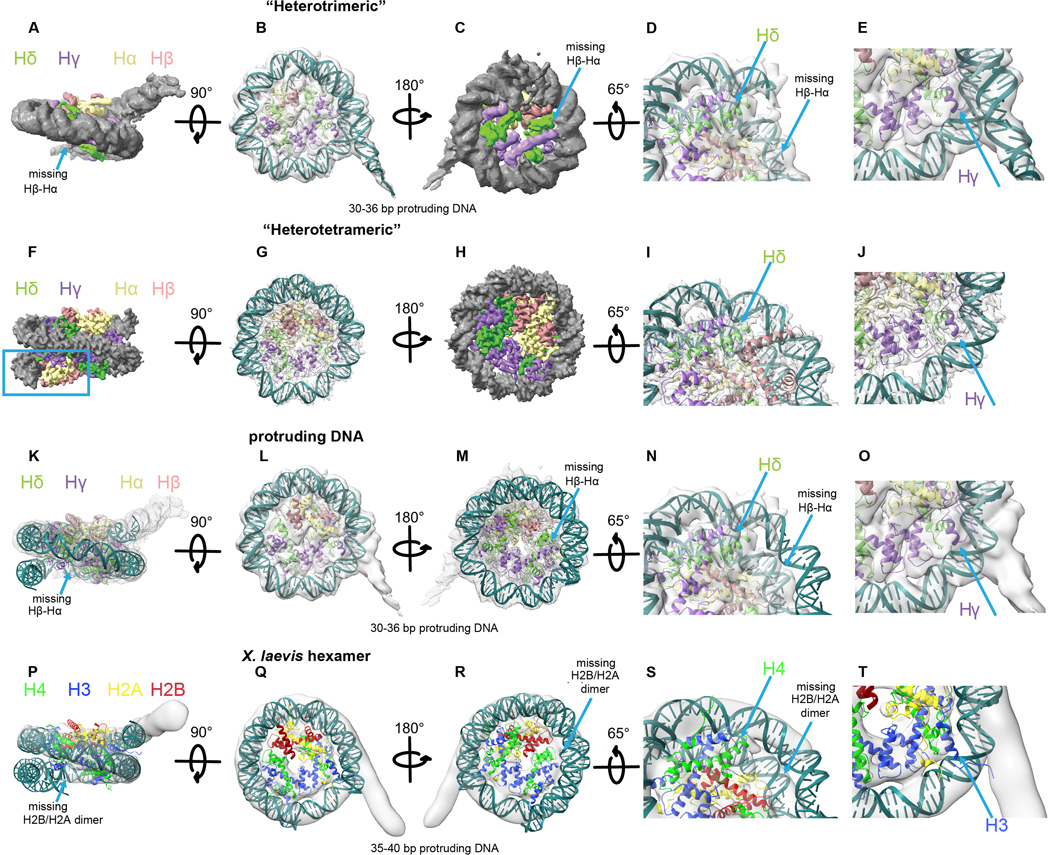
Extended Data Fig. 7. Msv “heterotrimeric” nucleosome.
a-e, Model of the “heterotrimeric” Msv nucleosome with modeled DNA fitting inside the “heterotrimeric” Msv cryo-EM map. a-c, Three different views, d, close up of the region where DNA is missing and e, close-up of the region where DNA is protruding. f-j, Model of the “heterotetrameric” Msv nucleosome fitting inside the “heterotetrameric” Msv cryo-EM map. f-h, Three different views, i and j, close ups of the regions shown in d and e. k-o, Model of the “heterotrimeric” Msv nucleosome without the modeled DNA fitting inside the “heterotrimeric” Msv cryo-EM map. k-m, Three different views, n and o, close ups of the regions shown in d and e. p-t, Docking of a model of a “hexameric” eukaryotic nucleosome (PDB ID 5AV9) produced taking off one of the dimers H2B and H2A and fitted inside the “hexameric” Xenopus cryo-EM map EMD-3929. Please note that we did not rebuilt the DNA in this model. p-r, Three different views, s and t,. close ups of the regions shown in d and e. Maps and models are color coded as Fig. 1. The blue square shows the missing DNA region and the arrow the missing Hβ-Hα (or H2B and H2A) histones.