Fig 10. Effect of dTat-gK4 peptide on latency and exhaustion marker levels in C57BL/6 infected mice.
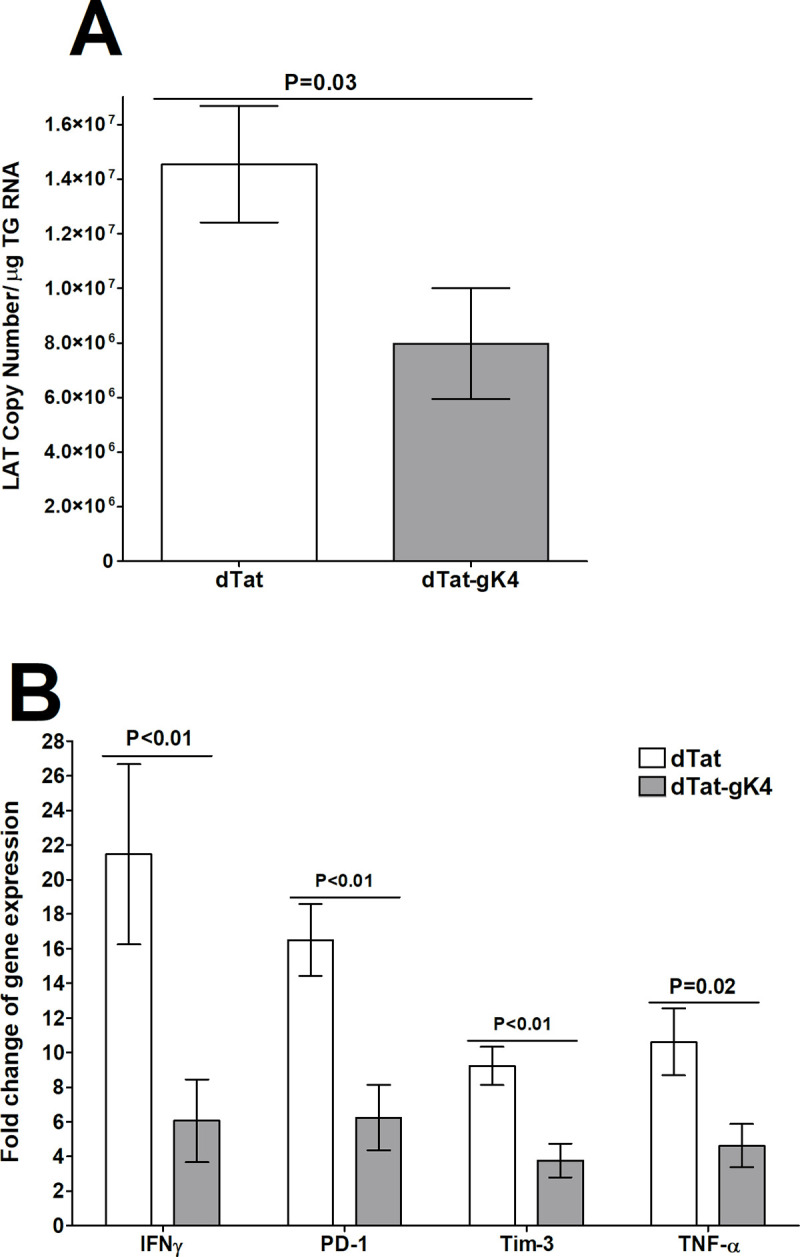
TG from mice described in Fig 9 legend were harvested on day 28 PI. Total RNA from each TG was isolated as described in Materials and Methods. A) Quantitation of LAT RNA in TG of immunized mice. Quantitative RT-PCR was performed on each mouse TG. In each experiment, an estimated relative copy number of HSV-1 LAT was calculated using standard curves generated from pGem5317. Briefly, DNA template was serially diluted 10-fold such that 1 μl contained 103 to 1011 copies of LAT, then subjected to TaqMan PCR with the same primer set. By comparing the normalized threshold cycle (CT) of each sample to the threshold cycle of the standard, copy number for each reaction was determined. GAPDH expression was used to normalize relative expression of viral LAT RNA in TG. Each bar represents the mean ± SEM from 22 TG for dTat-gK4 treated mice and 18 TG for dTat control group. B) qRT-PCR analyses of IFNγ, PD-1, Tim-3, and TNF-α transcripts in TG of latently-infected mice. Total RNA described above from each individual TG was used to estimate relative expression of IFNγ, PD-1, Tim-3, and TNF-α transcripts in TG of mice in dTat-gK4 treated or dTat control group. The expression ratio of each mRNA in the TG was normalized to its expression in TG of uninfected control mice. GAPDH expression was used to normalize relative expression of each transcript in TG of immunized mice. Each bar represents the mean ± SEM from 12 TG for each group. Panels: A) LAT; and B) Exhaustion markers.
