Fig. 3 |. DNA methylation stochasticity relates to gene expression in ETV6-RUNX1 ALL.
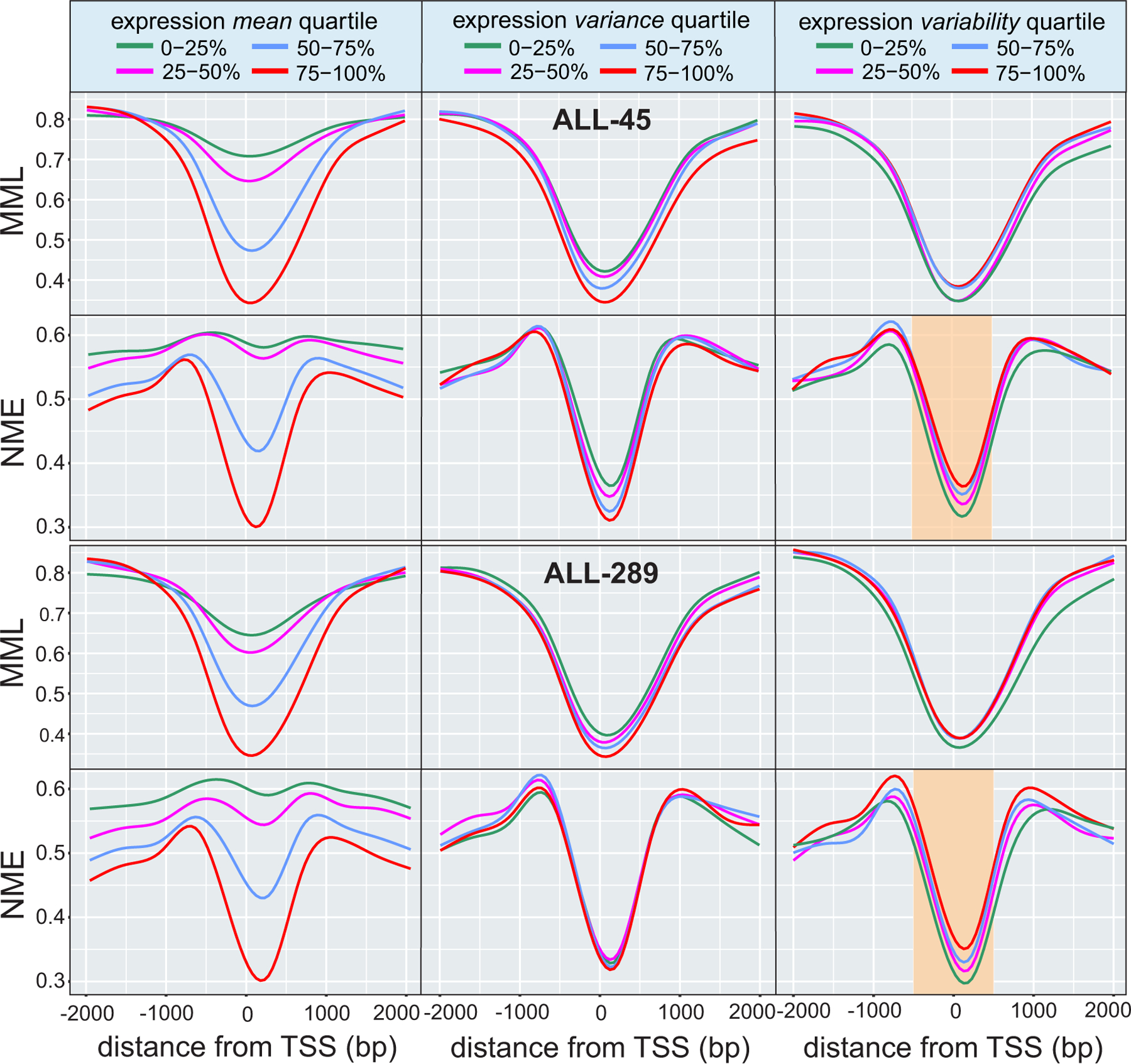
Average relationships in ETV6-RUNX1 ALL between mean methylation level (MML) and normalized methylation entropy (NME) within gene promoters as a function of distance from the transcription start site (TSS). The results correspond to quartiles of gene expression mean (left column), variance (middle column), and variability level (right column). Lower mean expression associates with higher levels of mean methylation (left column, first and third rows), confirming a known relationship between promoter methylation and gene expression. However, lower levels of mean expression are associated with higher levels of normalized methylation entropy (left column, second and fourth rows) implying that promoters of genes with lower gene expression are associated with higher levels of methylation stochasticity. Higher expression variance also relates to reduced levels of mean methylation level and normalized methylation entropy, but these associations can be weak (center column). Although a measure of expression variability (Methods) does not clearly associate with mean methylation level (right column, first and third rows), it relates to normalized methylation entropy, with higher entropy near the TSS being identified with statistically significant gains in expression variability (right column, second and fourth rows - highlighted: two-sided Wilcoxon rank sum test on medians within from the TSS, P-values for second vs. first quartile, third vs. second quartile, and fourth vs. third quartile). This implies that promoter regions of genes with higher expression variability exhibit higher levels of methylation stochasticity near the TSS.
