Fig. 5 |. UHRF1 is a target of epigenetic disruption in ALL.
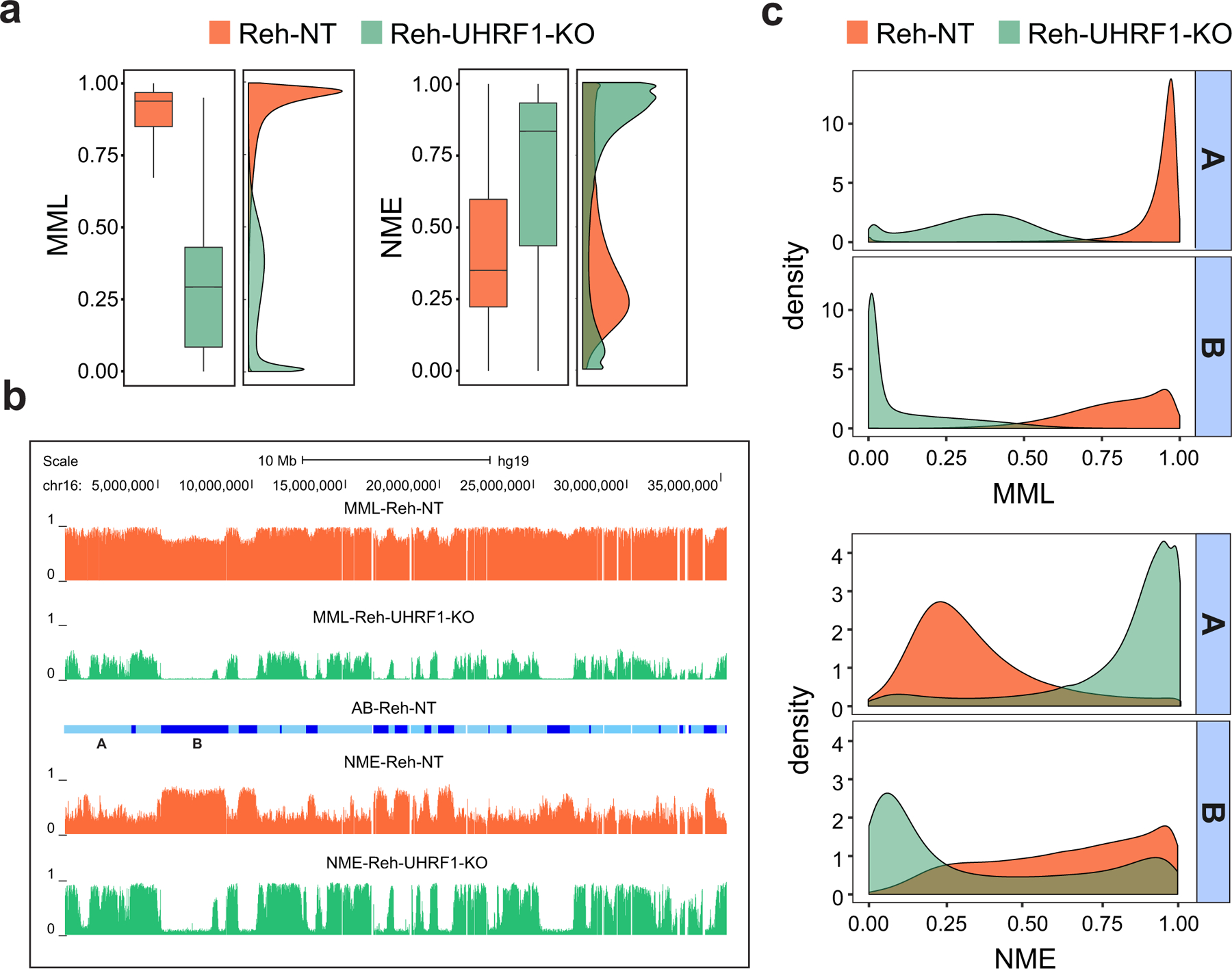
a, Boxplots and densities of genome-wide distributions of mean methylation level (MML) and normalized methylation entropy (NME) values in NT and UHRF1-KO WGBS samples show that UHRF1 silencing results in profound global hypomethylation and marked gain in normalized methylation entropy. Center lines, median; boxes, interquartile range (IQR); whiskers, 1.5 × IRQ. b, UCSC genome browser example showing that chromosome 16 exhibits profound hypomethylation and marked gain in normalized methylation entropy in UHRF1-KO Reh cells over NT associated euchromatic A domains (light blue), as well as almost zero mean methylation level and profound loss in normalized methylation entropy over heterochromatic B domains and over several genomic regions within A domains (dark blue). c, Densities of mean methylation level and normalized methylation entropy within NT associated A/B domains confirm the previous findings genome-wide.
