Abstract
Sense-antisense mRNA pairs generated by convergent transcription is a way of gene regulation. c-fms gene is closely juxtaposed to the HMGXB3 gene in the opposite orientation, in chromosome 5. The intergenic region (IR) between c-fms and HMGXB3 genes is 162 bp. We found that a small portion (∼4.18%) of HMGXB3 mRNA is transcribed further downstream, including the end of the c-fms gene generating antisense mRNA against c-fms mRNA. Similarly, a small portion (∼1.1%) of c-fms mRNA is transcribed further downstream, including the end of the HMGXB3 gene generating antisense mRNA against the HMGXB3 mRNA. Insertion of the strong poly(A) signal sequence in the IR results in decreased c-fms and HMGXB3 antisense mRNAs, resulting in up-regulation of both c-fms and HMGXB3 mRNA expression. miR-324-5p targets HMGXB3 mRNA 3′ UTR, and as a result, regulates c-fms mRNA expression. HuR stabilizes c-fms mRNA, and as a result, down-regulates HMGXB3 mRNA expression. UALCAN analysis indicates that the expression pattern between c-fms and HMGXB3 proteins are opposite in vivo in breast cancer tissues. Together, our results indicate that the mRNA encoded by the HMGXB3 gene can influence the expression of adjacent c-fms mRNA, or vice versa.
Keywords: convergent transcription termination, sense-antisense RNA pairing, mRNA 3′ end polymorphism, c-fms mRNA 3′ end, HMGXB3 mRNA 3′ end
INTRODUCTION
In RNA polymerase II (RNAPII)-driven transcription elongation, termination, transcript cleavage, and release commonly occur after recognizing the poly(A) site (PAS) by the 3′ end cleavage and polyadenylation (CPA) complex (West et al. 2008; Kuehner et al. 2011; Proudfoot 2016). Cleavage by CPA releases the nascent transcript, which becomes polyadenylated at the 3′ end (Proudfoot 1989). When more than one PAS is present, alternative polyadenylation occurs.
Transcription termination can also occur anywhere from the 3′ end of the mRNA by readthrough transcription (Proudfoot 1989; Dye and Proudfoot 1999; Richard and Manley 2009), which generates the mRNA 3′ end polymorphism (de Klerk and ‘t Hoen 2015; Kainov et al. 2016; Nourse et al. 2020). In yeast, various transcript boundaries are present (Gullerova and Proudfoot 2008; Pelechano et al. 2013).
In the case of a convergent gene pair, RNAPII molecules in the transcription elongation complex collide head-to-head and stop transcription (Hobson et al. 2012). In head-to-head collision, RNAPII molecules may also bypass one another, generating sense-antisense transcript pairs overlapping at the 3′ end (Ma and McAllister 2009). In humans, over 20% of transcripts may form sense-antisense pairs, in which most antisense transcripts (i.e., cis-encoded natural antisense, cis-NAT) are generated from the opposite strand of the same genomic locus of the sense strand (Yelin et al. 2003; Chen et al. 2004; Zhang et al. 2006).
Natural antisense transcripts (NATs) are prevalent in human cells (Zhang et al. 2006; Pelechano and Steinmetz 2013). cis-NATs have perfect sequence complementary to the sense transcripts. cis-NATs can be derived from tail-to-tail (3′ to 3′) to their relative orientation (Lapidot and Pilpel 2006). NATs are proposed to be involved in several regulatory mechanisms. NAT can interfere with sense transcription by collision of two RNAPII complexes. NAT can also mask sense mRNA and hinder protein–RNA interaction in the cytoplasm, resulting in translation inhibition and RNA decay. Interaction of 3′ overlapping mRNAs promotes no-go-decay in the cytoplasm (Sinturel et al. 2015).
In RNA interference (RNAi), dsRNA formed by convergent transcription is processed to generate siRNAs, which are incorporated into the RNAi-induced silencing (RISC) complex for posttranscriptional gene silencing (PTGS) by inducing mRNA decay and translational repression (Borsani et al. 2005; Kim and Nam 2006; Carthew and Sontheimer 2009; Pelechano and Steinmetz 2013). In the nucleus, siRNA derived from dsRNA by convergent transcription is involved in transcriptional gene silencing (TGS) (Gullerova and Proudfoot 2012). In TGS, siRNA-induced transcriptional silencing (RITS) complex targets homologous gene loci, which in turn induce heterochromatin formation resulting in gene silencing (Buhler and Moazed 2007). In this aspect, TGS and PTGS are proposed to cooperate for gene silencing (Faghihi and Wahlestead 2009; Werner and Sayer 2009).
Aberrant mRNA 3′ end cleavage and processing are known to associate with diseases including cancer (Danckwardt et al. 2008; Di Giammartino et al. 2011; Ogorodnikov et al. 2016; Nourse et al. 2020). Both point mutations in PAS as well as mutations in CPA complexes have resulted in aberrant 3′ end processing and pathogenesis.
Here, we describe gene regulation by cis-NATs generated from a convergent gene pair consisting of the proto-oncogene c-fms and the HMGXB3 gene. The proto-oncogene c-fms, which encodes the receptor tyrosine kinase and a sole receptor for CSF-1, is expressed by the tumor epithelium in several human epithelial cancers (Kacinski et al. 1988, 1990, 1991). An elevated level of c-fms is associated with poor prognosis (Chambers et al. 1997; Maher et al. 1998). Activated (phosphorylated) c-fms protein in breast and ovarian cancer is present in over 50% of tumors (Flick et al. 1997; Toy et al. 2001). In breast cancer, interaction of c-fms bearing tumor associated macrophages in breast tissues promotes malignant transformation (Lin et al. 2001). HuR is a member of the Elav/Hu family of RNA-binding proteins and plays a supportive role for c-fms in breast cancer progression by binding a pyrimidine-rich sequence in its mRNA 3′ UTR, thus regulating its expression (Woo et al. 2009).
HMGXB3 belongs to a high-mobility group binding protein 3 family. Currently, there is limited information about HMGXB3 at the molecular level. However, HMGXB3 is known to be involved in cell proliferation and migration (Guo et al. 2016; Sing et al. 2019). Knockdown of HMGXB3 inhibits cell proliferation and reduces migration in gastric cancer cells (Guo et al. 2016) and non-small cell lung cancer cells (Song et al. 2019). miR-324-5p targets HMGXB3 mRNA 3′ UTR and down-regulates its expression (Sun et al. 2017).
We report here that the proto-oncogene c-fms and HMGXB3 are a convergent gene pair with a 162 bp intergenic region between their 3′ ends. They each generate mRNAs with extended 3′ ends which are cis-antisense RNA against the 3′ end of their pair mRNA. Expression of both genes regulate each other by sense-antisense RNA pairing, which may be self-regulatory circuits to regulate their own expression.
RESULTS
c-fms and HMGXB3 mRNA 3′ end polymorphisms are derived from extended transcription termination
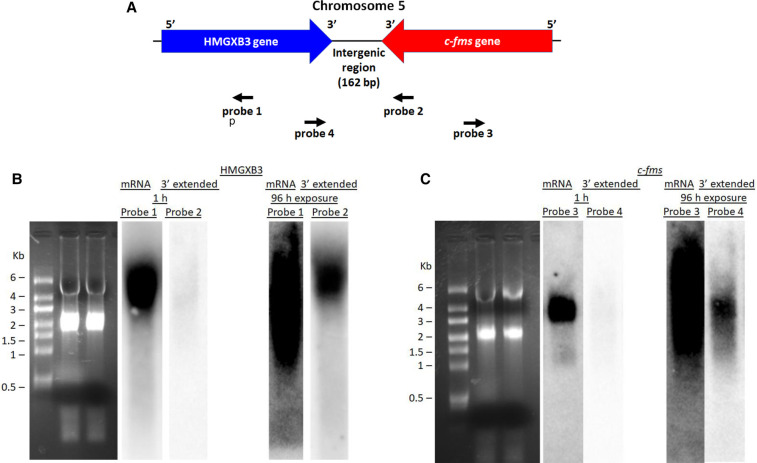
Both c-fms and HMGXB3 genes are closely juxtaposed in chromosome 5. The intergenic region between two genes is 162 bp (Fig. 1A). Since both genes are closely located in the opposite orientation, we checked the presence of mRNA 3′ end extension generated by readthrough convergent transcription termination. For northern blot analysis, strand-specific antisense RNA probes 1 through 4 were generated to detect either c-fms or HMGXB3 mRNA with 3′ end extension (Supplemental Fig. S1). Northern blot analysis indicates that HMGXB3 mRNA is detected by strand-specific antisense RNA probe 1 generated from the HMGXB3 mRNA coding region in 1 h exposure (Fig. 1B). In addition, HMGXB3 mRNA with 3′ end extension is also detected by strand-specific antisense RNA probe 2 generated from the c-fms mRNA 3′ UTR in 96 h exposure (Fig. 1B). Strand-specific antisense RNA probe 2 cannot detect c-fms mRNA, since it is sense c-fms RNA.
FIGURE 1.
c-fms and HMGXB3 mRNA 3′ end polymorphism. (A) Both HMGXB3 gene and c-fms gene are closely juxtaposed in chromosome 5. The intergenic region is 162 bp. 32P-labeled strand-specific antisense RNA probe 1 derived from the HMGXB3 mRNA coding region detects HMGXB3 mRNA. 32P-labeled strand-specific antisense RNA probe 2 derived from the 3′ end of c-fms mRNA detects 3′ end extended HMGXB3 mRNA. 32P-labeled strand-specific antisense RNA probe 3 derived from c-fms mRNA coding region detects c-fms mRNA. 32P-labeled strand-specific antisense RNA probe 4 derived from the 3′ end of HMGXB3 mRNA detects 3′ end extended c-fms mRNA. Detailed description of probe generation is in Supplemental Figure S1. (B) Northern blot shows HMGXB3 mRNA and HMGXB3 mRNA with 3′ end extension. HMGXB3 mRNA is detected in 1 h exposure. In contrast, HMGXB3 mRNA with 3′ end extension is detected in 96 h exposure. (C) Northern blot shows c-fms mRNA and c-fms mRNA with 3′ end extension. C-fms mRNA is detected in 1 h exposure. In contrast, c-fms mRNA with 3′ end extension is detected in 96 h exposure.
Northern blot analysis also indicates that c-fms mRNA is detected by strand-specific antisense RNA probe 3 generated from the c-fms mRNA coding region in 1 h exposure (Fig. 1C). In addition, c-fms mRNA with 3′ end extension is also detected by strand-specific antisense RNA probe 4 generated from the HMGXB3 mRNA 3′ UTR in 96 h exposure (Fig. 1C). Strand-specific antisense RNA probe 4 cannot detect HMGXB3 mRNA, since it is sense HMGXB3 RNA.
This indicates that a small portion of c-fms and HMGXB3 mRNAs with 3′ end extensions are present, since weak signals are detected by strand-specific antisense RNA probes 2 and 4 in 96 h exposure.
Mapping and quantification of c-fms and HMGXB3 mRNAs with 3′ end extensions
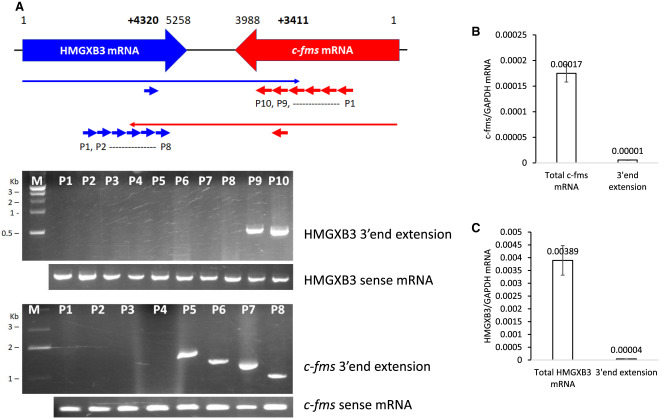
To map the 3′ end extensions of c-fms and HMGXB3 mRNAs, we did RT-PCR with strand-specific primers and poly-(A)+-RNA (Fig. 2A). Reverse transcription was performed with strand-specific primers from multiple different locations of c-fms and HMGXB3 mRNAs (Fig. 2A; Supplemental Fig. S2A,B; Supplemental Table S1). The 3′ end of HMGXB3 mRNA is extended up to 578 nt inside of the c-fms mRNA 3′ UTR (i.e., up to c-fms mRNA +3411nt). The 3′ end of c-fms mRNA is extended up to 939 nt inside of the HMGXB3 mRNA 3′ UTR (i.e., up to HMGXB3 mRNA +4320 nt).
FIGURE 2.
Mapping and quantification of transcripts with the 3′ end extensions. (A) Mapping of 3′ end extension by RT-PCR. The HMGXB3 mRNA with the 3′ end extension was reverse transcribed by 10 primers derived from the c-fms mRNA sequence. RT transcript was PCR amplified for mapping. The c-fms mRNA with the 3′ end extension was reverse transcribed by eight primers derived from the HMGXB3 mRNA sequence. RT transcript was PCR amplified for mapping. A detailed mapping strategy is described in Supplemental Figure S2. (B) qRT-PCR indicates that c-fms mRNA with 3′ end extension is 4.1% of total c-fms mRNA (n = 4). (C) qRT-PCR indicates that HMGXB3 mRNA with 3′ end extension is 1.1% of total HMGXB3 mRNA (n = 4). Detailed quantification strategy is described in Supplemental Figure S2.
For further quantification, we measured c-fms and HMGXB3 total mRNAs and also 3′ end extensions by strand-specific qRT-PCR (described in Supplemental Fig. S2C,D). Quantification of both mRNAs and the 3′ end extensions indicates that c-fms mRNA with the 3′ end extension is ∼4.1% of total c-fms mRNA (Fig. 2B; Supplemental Fig. S2E). Similarly, HMGXB3 mRNA with the 3′ end extension is ∼1.1% of total HMGXB3 mRNA (Fig. 2C; Supplemental Fig. S2F).
We conclude that a small portion of c-fms and HMGXB3 mRNAs with the 3′ end extensions are generated by readthrough convergent transcription termination. These c-fms and HMGXB3 mRNAs with the 3′ end extensions also have poly-(A)+ tails, since poly-(A)+-RNA was used for mapping (Fig. 2A).
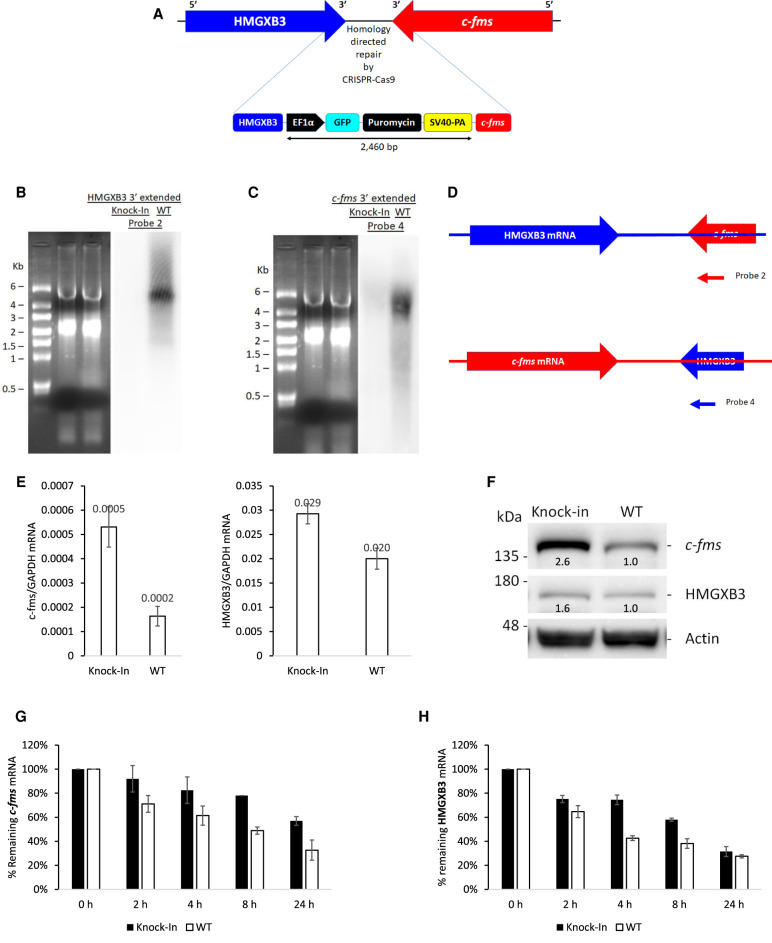
Introduction of strong poly(A) signal sequence in the intergenic region increases both c-fms and HMGXB3 expression
To block the extended transcription termination, a strong poly(A) signal sequence SV40-PA (Schek et al. 1992; Hans and Alwine 2000) was introduced in the intergenic region (IR) in chromosome 5 via CRISPR/Cas9 (Fig. 3A). Insertion of SV40-PA was confirmed by genome PCR and sequencing (Supplemental Fig. S3A). Insertion of SV40-PA in the IR abolishes the expression of HMGXB3 mRNA with the 3′ end extension (which is antisense to the c-fms mRNA 3′ end) (Fig. 3B Northern) thereby increasing the steady-state level of c-fms mRNA by 2.5-fold (n = 4, Fig. 3E; Supplemental Fig. S3B) and protein by 2.6-fold (Fig. 3F) compared to the wild-type cells. Insertion of SV40-PA in the IR also abolishes the expression of c-fms mRNA with the 3′ end extension (which is antisense to the HMGXB3 mRNA 3′ end) (Fig. 3C Northern) thereby increasing the steady-state level of HMGXB3 mRNA by 1.46-fold (n = 4, P < 0.001, Fig. 3E; Supplemental Fig. S3C) and protein by 1.6-fold (±0.32, n = 4, Fig. 3F; Supplemental Fig. S3E).
FIGURE 3.
Insertion of strong poly(A) signal in the intergenic region increases the expression of both c-fms and HMGXB3. (A) SV40-PA is inserted in the intergenic region by CRISPR–Cas9. Confirmation of insertion is shown in Supplemental Figure S3. (B) SV40-PA knock-in abolishes the expression of HMGXB3 mRNA with 3′ end extension, and (C) the expression of c-fms mRNA with 3′ end extension. Northern blot was exposed for 96 h. (D) 32P-labeled strand-specific antisense probe 2 is derived from the c-fms mRNA 3′ end. 32P-labeled strand-specific antisense probe 4 is derived from HMGXB3 mRNA 3′ end. 32P-labeled strand-specific probes 2 and 4 were synthesized as described in Supplemental Figure S1. (E) SV40-PA knock-in increases c-fms mRNA and HMGXB3 mRNA (n = 4). (F) SV40-PA knock-in increases both c-fms protein and HMGXB3 protein. Numbers below bands indicate band intensities scanned by ImageJ. (G) SV40-PA knock-in increases c-fms mRNA half-life, and (H) HMGXB3 mRNA half-life (n = 3).
When the wild-type (WT) group in BT20 cells was compared with the SV40-PA inserted group, the half-life of c-fms mRNA increased from 8 h to >24 h (two-tailed t-test, n = 3, P < 0.001) (Fig. 3G). The half-life of HMGXB3 mRNA also increased from 4 h to >8 h (two-tailed t-test, n = 3, P < 0.001) (Fig. 3H).
These results underscore the stabilizing influence of SV40-PA insertion by down-regulating c-fms or HMGXB3 mRNAs with the 3′ end extensions, which serve as natural antisense RNAs.
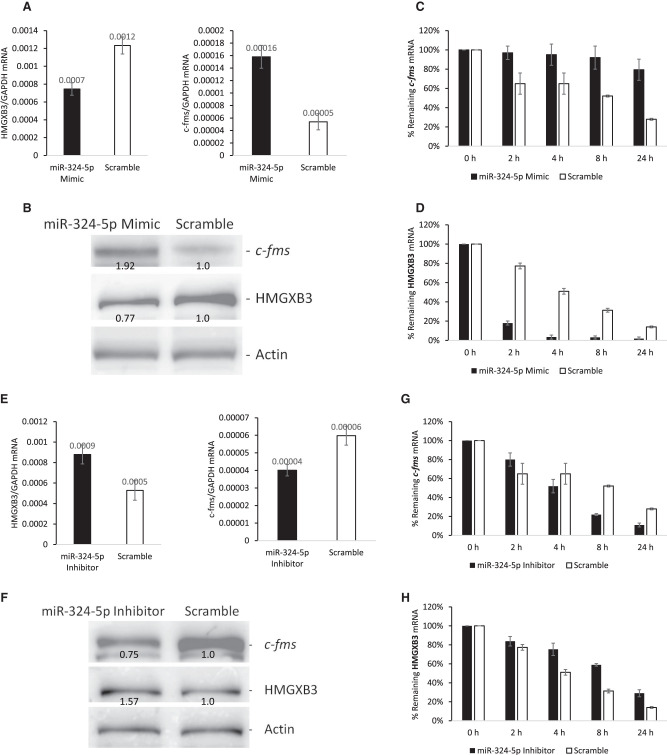
miR-324-5p regulates c-fms mRNA expression via interacting with HMGXB3 mRNA
Sun et al. (2017) reported that miR-324-5p interacts with HMGXB3 mRNA 3′ UTR. TargetScan (http://www.targetscan.org/vert 72/) for prediction of miRNA targets also predicts the interaction of miR-324-5p with HMGXB3 mRNA, but not with c-fms mRNA. We checked whether the interaction of miR-324-5p with HMGXB3 mRNA influences the expression of c-fms mRNA. miR-324-5p mimic down-regulates the steady-state level of HMGXB3 mRNA and up-regulates c-fms mRNA (n = 4, Fig. 4A; Supplemental Fig. S4A,B). A similar effect was also observed in their protein levels, that is, miR-324-5p mimic down-regulates HMGXB3 protein expression and up-regulates c-fms protein expression (Fig. 4B; Supplemental Fig. S4C).
FIGURE 4.
miR-324-5p regulates c-fms mRNA expression thru HMGXB3 mRNA. (A) miR-324-5p mimic decreases HMGXB3 mRNA and increases c-fms mRNA (n = 4). (B) Western analysis shows increased expression of c-fms protein and decreased expression of HMGXB3 protein by miR-324-5p mimic. Numbers below bands indicate band intensities scanned by ImageJ. (C) miR-324-5p mimic increases c-fms mRNA half-life (n = 3). (D) miR-324-5p mimic decreases HMGXB3 mRNA half-life (n = 3). (E) miR-324-5p inhibitor increases HMGXB3 mRNA and decreases c-fms mRNA (n = 4). (F) Western analysis shows decreased expression of c-fms protein and increased expression of HMGXB3 protein by miR-324-5p inhibitor. Numbers below bands indicate band intensities scanned by ImageJ. (G) miR-324-5p inhibitor decreases c-fms mRNA half-life (n = 3). (H) miR-324-5p inhibitor increases HMGXB3 mRNA half-life (n = 3).
We studied the effects of altering HMGXB3 mRNA levels by miR-324-5p mimic on c-fms mRNA half-life in BT20 cells. As shown in Figure 2C, at least part of the 3.2-fold increase in the steady-state level of c-fms mRNA in BT20 cells seen on miR-324-5p mimic (Fig. 4A) is due to an increase in the stability of c-fms mRNA (Fig. 4C). When the scramble control group in BT20 cells was compared with the miR-324-5p mimic group, the half-life of c-fms mRNA increased from 8 h to >24 h (two-tailed t-test, n = 3, P < 0.001). These results underscore the stabilizing influence of miR-324-5p mimic on c-fms mRNA. At the same time, the half-life of HMGXB3 mRNA decreased from 4 h to <2 h (two-tailed t-test, n = 3, P < 0.001). These results also underscore the destabilizing influence of miR-324-5p mimic on HMGXB3 mRNA (Fig. 4D).
Conversely, miR-324-5p inhibitor up-regulates the steady-state level of HMGXB3 mRNA, and down-regulates c-fms mRNA (n = 4, Fig. 4E; Supplemental Fig. S4D,E). A similar effect was also observed in protein levels, that is, miR-324-5p inhibitor up-regulates HMGXB3 protein expression and down-regulates c-fms protein expression (Fig. 4F; Supplemental Fig. S4F).
We also studied the effects of altering HMGXB3 mRNA level by miR-324-5p inhibitor on c-fms mRNA half-life in BT20 cells. When the scramble control group in BT20 cells was compared with the miR-324-5p inhibitor group, the half-life of c-fms mRNA decreased from 8 h to ∼4 h (two-tailed t-test, n = 3, P < 0.001) (Fig. 4G). These results underscore the destabilizing influence of miR-324-5p inhibitor on c-fms mRNA. At the same time, the half-life of HMGXB3 mRNA increased from 4 h to >8 h (two-tailed t-test, n = 3, P < 0.001) (Fig. 4H). These results also underscore the stabilizing influence of miR-324-5p inhibitor on HMGXB3 mRNA.
We conclude that miR-324-5p regulates c-fms mRNA expression via interacting with HMGXB3 mRNA.
miR-324-5p targets HMGXB3 mRNA 3′ UTR, not c-fms mRNA 3′ UTR
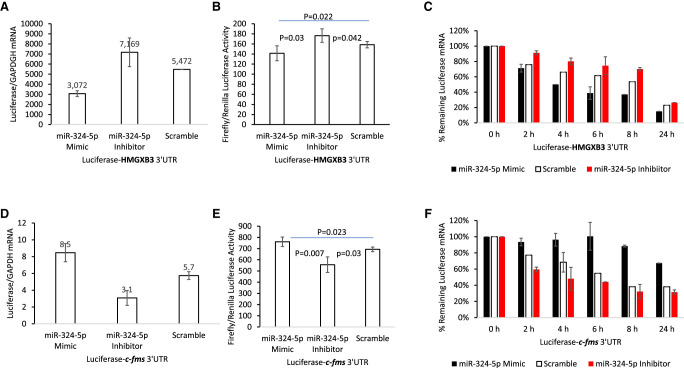
Since Sun et al. (2017) reported direct targeting of miR-324-5p to HMGXB3 mRNA 3′ UTR, we further verified that miR-324-5p does not also target the 3′ UTR of c-fms mRNA. To do this, we made luciferase reporter fused with either HMGXB3 or c-fms mRNA 3′ UTRs (Supplemental Fig. S5A). BT20 cells were cotransfected either miR-324-5p mimic or miR-324-5p inhibitor with luciferase-3′ UTR reporter plasmids bearing 3′ UTR sequence of either c-fms or HMGXB3 mRNA. As expected, miR-324-5p mimic reduced the steady-state luciferase mRNA and activity of HMGXB3 reporter plasmid (n = 4, Fig. 5A,B; Supplemental Fig. S5B). Conversely, the steady-state luciferase-HMGXB3 mRNA 3′ UTR and activity were increased by cotransfected with miR-324-5p inhibitor.
FIGURE 5.
Luciferase reporter assay indicates that miR-324-5p has opposite effects on HMGXB3 and c-fms expression. (A,B) miR-324-5p mimic decreases and miR-324-5p inhibitor increases luciferase-HMGXB3 mRNA 3′ UTR reporter expression (n = 4). (C) miR-324-5p mimic decreases (black bar) and miR-324-5p inhibitor increases luciferase-HMGXB3 mRNA 3′ UTR reporter RNA half-life (red bar) (n = 3). (D,E) miR-324-5p mimic increases and miR-324-5p inhibitor decreases luciferase-c-fms mRNA 3′ UTR reporter expression (n = 4). (F) miR-324-5p mimic increases (black bar) and miR-324-5p inhibitor decreases luciferase-c-fms mRNA 3′ UTR reporter RNA half-life (red bar) (n = 3).
As shown in Figure 5C, at least part of the 2.5-fold decrease in the steady-state level of luciferase-HMGXB3 mRNA in BT20 cells seen in the miR-324-5p mimic group (Fig. 5A) is due to a decrease in the stability of luciferase-HMGXB3 mRNA (Fig. 5C). When the scramble control group in BT20 cells was compared with the miR-324-5p mimic group, the half-life of luciferase-HMGXB3 mRNA 3′ UTR decreased from 6 h to 4 h (two-tailed t-test, n = 3, P < 0.001). These results underscore the destabilizing influence of miR-324-5p mimic on luciferase-HMGXB3 mRNA 3′ UTR. At the same time, the half-life of luciferase-HMGXB3 mRNA 3′ UTR increased from 6 h to >8 h (two-tailed t-test, n = 3, P < 0.001) in the miR-324-5p inhibitor treatment group. These results also underscore the stabilizing influence of miR-324-5p inhibitor on luciferase-HMGXB3 mRNA 3′ UTR (Fig. 5C).
In contrast, miR-324-5p mimic increased the steady-state level of luciferase mRNA and activity of c-fms mRNA 3′ UTR reporter plasmid (n = 4, Fig. 5D,E; Supplemental Fig. S5C). This increase is likely due to the decrease of the HMGXB3 mRNA with 3′ end extension (which is antisense to the c-fms mRNA 3′ end in luciferase construct). Conversely, the steady-state luciferase-c-fms mRNA 3′ UTR and activity were decreased by cotransfection with miR-324-5p inhibitor. This decrease is also likely due to the increase of the HMGXB3 mRNA with 3′ end extension. When the scramble control group in BT20 cells was compared with the miR-324-5p mimic group, the half-life of luciferase-c-fms mRNA 3′ UTR increased from 6 h to >24 h (two-tailed t-test, n = 3, P < 0.001) (Fig. 5F). These results underscore the destabilizing influence of miR-324-5p mimic on the HMGXB3 3′ end extended transcript. At the same time, the half-life of luciferase-c-fms mRNA 3′ UTR decreased from 6 h to 4 h (two-tailed t-test, n = 3, P < 0.001) in the miR-324-5p inhibitor treatment group. These results also underscore the stabilizing influence of miR-324-5p inhibitor on the HMGXB3 mRNA with 3′ end extension.
We conclude that miR-324-5p directly targets HMGXB3 mRNA 3′ UTR, not the c-fms mRNA 3′ UTR.
HuR up-regulates c-fms mRNA and down-regulates HMGXB3 mRNA expression
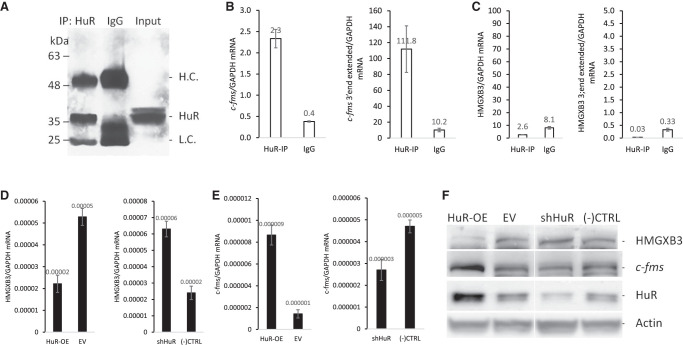
We previously reported that HuR, an RNA binding protein, interacts and stabilizes c-fms mRNA, resulting in an increase of c-fms expression (Woo et al. 2009). To test whether HuR also directly associates with HMGXB3 mRNA, IP assay was performed in BT20 cells (Fig. 6A). Possible association of HMGXB3 mRNA with HuR was determined by isolating RNA from the IP material and analyzing it by quantitative real-time PCR. As shown in Figure 6B, in cellular lysates, the c-fms mRNA was dramatically enriched in HuR IP samples compared to that in control IgG IP samples. The association of c-fms mRNA with HuR was 5.8-fold higher than that seen in the control IgG IP reaction (Fig. 6B, n = 3). In contrast, the association of HMGXB3 mRNA with HuR was not observed (Fig. 6C, n = 3), indicating HuR does not directly associate with HMGXB3 mRNA.
FIGURE 6.
HuR targets c-fms mRNA, not HMGXB3 mRNA. (A) HuR IP of BT20 cell lysates. Two-fold excess IgG was used for nonspecific binding. (B) HuR IP enriches c-fms mRNA and c-fms mRNA with the 3′ end extension (n = 3), (C) not HMGXB3 mRNA and HMGXB3 mRNA with the 3′ end extension (n = 3). (D) HuR overexpression decreases and shHuR increases HMGXB3 mRNA (n = 4). (E) HuR overexpression increases and shHuR decreases c-fms mRNA (n = 4). (F) Western analysis shows that HuR overexpression decreases and shHuR increases HMGXB3 protein. In contrast, HuR overexpression increases and shHuR decreases c-fms protein.
Since HuR stabilizes c-fms mRNA (Woo et al. 2009), we checked whether c-fms mRNA stabilization down-regulates HMGXB3 expression. HuR overexpression down-regulates HMGXB3 expression (Fig. 6D–F; Supplemental Fig. S6A–F, n = 4). As expected, c-fms expression is up-regulated by HuR overexpression. In contrast, down-regulation of HuR by shRNA up-regulates HMGXB3 expression and down-regulates c-fms expression.
We conclude that HuR stabilizes both c-fms mRNA and c-fms mRNA with 3′ end extension (which is antisense to HMGXB3 mRNA 3′ end), and as a result, down-regulates HMGXB3 mRNA expression.
The expression pattern is opposite between c-fms and HMGXB3 proteins in breast cancer
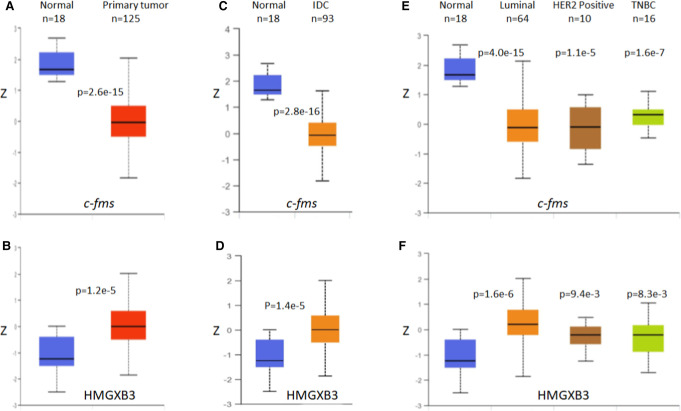
UALCAN data set (http://ualcan.path.uab.edu/) (Chandrashekar et al. 2017), a comprehensive web resource for analyzing cancer OMICS data, was utilized to identify the protein levels of c-fms and HMGXB3 between breast cancer tissues and normal tissues. The UALCAN data analyses indicate that the expression pattern between c-fms and HMGXB3 proteins is opposite, that is, the c-fms protein level is lower, and the HMGXB3 protein level is higher in general in breast cancer tissues than normal breast tissue (Fig. 7A,B). HMGXB3 is up-regulated and c-fms is down-regulated in samples with infiltrating ductal carcinoma (Fig. 7C,D). Luminal, her2/neu, or triple negative breast cancers (TNBC) all showed high expression of HMGXB3 and low expression of c-fms compared with normal breast cancer tissue (Fig. 7E,F). This analysis indicates that the expression pattern is opposite between the two proteins in vivo and supports our in vitro findings.
FIGURE 7.
The protein expression of c-fms and HMGXB3 in breast cancer (UALCAN). (A) The expression of c-fms protein in breast cancer tissues is lower than that in normal tissues. (B) In contrast, the expression of HMGXB3 protein in breast cancer tissues is higher than that in normal tissues. (C,D) Similar trends are shown in histologic subtypes, and (E,F) breast cancer subclasses. Z-values on the y-axis represent standard deviations from the median across samples for the given cancer type. Log2 spectral count ratio values from CPTAC were first normalized within each sample profile, then normalized across samples. P-value less than 0.05 indicates significant differences between samples. (IDC) Infiltrating ductal carcinoma, (TNBC) triple negative breast cancer.
DISCUSSION
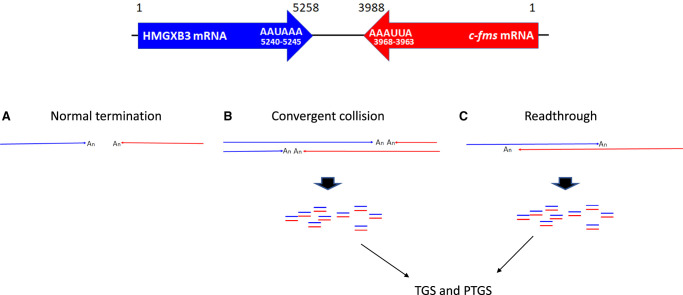
Transcription of the convergent gene pair generates mRNA 3′ end polymorphism by RNAPII collision or readthrough transcription (Fig. 8; Proudfoot 1989; Dye and Proudfoot 1999; Gullerova and Proudfoot 2008; Richard and Manley 2009; Pelechano et al. 2013; de Klerk and ‘t Hoen 2015; Kainov et al. 2016). The sense-antisense RNA pair generated by convergent transcription can modulate the expression of mRNA on a posttranscriptional level (PTGS) (Borsani et al. 2005; Kim and Nam 2006; Carthew and Sontheimer 2009; Pelechano and Steinmetz 2013) as well as transcriptional level (TGS) by establishing a local epigenetic imprint (Buhler and Moazed 2007; Gullerova and Proudfoot 2012). NAT generated by convergent transcription also masks sense mRNA and hinders protein–RNA interaction resulting in translation inhibition and RNA decay (Lapidot and Pilpel 2006; Zhang et al. 2006; Pelechano and Steinmetz 2013). The c-fms and HMGXB3 convergent gene pair generates mRNA 3′ end polymorphism (Figs. 1, 2) resulting in sense-antisense RNA pairing, which regulates their own expression (Figs. 3–6).
FIGURE 8.
Termination of convergent transcription elongation. (A) Transcription elongation complexes collide in the convergent gene pairs and terminate transcription. (B) Differential rate of transcription elongation between the convergent gene pairs results in the 3′ end polymorphism. (C) Transcriptional bypass also generates the 3′ end polymorphism. mRNA is depicted with the poly(An) tail. Poly(A) signal sequences are shown in HMGXB3 mRNA (5240–5245) and c-fms mRNA (3963–3968).
Even though both c-fms and HMGXB3 mRNAs have conserved poly(A) signal sequences which reside right upstream of the normal termination site, that is, AUUAAA (3963–3968) for c-fms mRNA and AAUAAA (5240–5245) for HMGXB3 mRNA (Fig. 8), a small portion of transcripts extends further downstream generating mRNAs with the 3′ end extensions, which serve as natural antisense RNAs (Figs. 1, 2). Furthermore, these c-fms and HMGXB3 mRNAs with the 3′ end extensions are still polyadenylated at the 3′ end (Fig. 2A). We do not find any specific sequence or structure for termination of extended transcription. We propose that RNAPII collision in convergent gene pair may randomly stop the transcription generating mRNA 3′ end polymorphism. The mechanism(s) are unknown at this time. There is a limit to RT-PCR mapping for determination of the exact transcription termination sites and the presented mapping (Fig. 2A) is the most representative. In the future, mRNA 3′ end transcriptome NGS needs to be done for accurate mapping and finding the population of variable ends of transcripts.
Knock-in of a strong poly(A) signal (SV40-PA) reduces bidirectional mRNA 3′ end extension, thereby reducing sense-antisense RNA pairing, and up-regulating both c-fms and HMGXB3 mRNA and protein expression (Fig. 3). The orientation of poly(A) signal sequence may also contribute to the efficiency of transcription termination. The SV40-PA is introduced in the 5′-to-3′ orientation to the end of HMGXB3 mRNA (Fig. 3), which results in dramatic down-regulation of HMGXB3 mRNA with the 3′ end extension (which is antisense RNA against c-fms mRNA) (Fig. 3B) resulting in a profound increase of c-fms mRNA (Fig. 3E) and protein (Fig. 3F). The extension of 3′ UTRs from both c-fms and HMGXB3 can cause a partial overlap in 3′ UTR regions, generating sense-antisense RNA pair resulting in either PTGS or TGS. The length of the overlapping region may be inversely correlated to the level of expression; that is, an increase of HMGXB3 mRNA (Fig. 3E) and protein (Fig. 3F) is not comparable with the dramatic increase of c-fms expression. Transcription termination of c-fms and HMGXB3 mRNAs may also occur at different time points or simultaneously in the cell.
The miR-324-5p and HuR studies indicate that the major effect of sense-antisense RNA pairs on reduction of mRNA and protein expression is largely on the basis of mRNA instability (Figs. 4–6). miR-324-5p and HuR effects are indirect on either c-fms or HMGXB3 mRNA expression, respectively. miR-324-5p down-regulates HMGXB3 mRNA and, as a result, increases c-fms mRNA (Fig. 4). In contrast, HuR up-regulates c-fms mRNA and, as a result, down-regulates HMGXB3 mRNA (Fig. 6).
Overexpression of c-fms mRNA and protein in breast cancer epithelium, including activated phosphorylated c-fms protein, has been observed both in vitro and in vivo (Kacinski et al. 1991; Sapi et al. 1995; Flick et al. 1997). Breast cancer tissues, analyzed in the CPTAC/TCGA projects (http://ualcan.path.uab.edu/) (Chandrashekar et al. 2017), comprise malignant breast epithelium and its surrounding stroma. In addition, the “normal” breast tissue is frequently collected adjacent to the tumor. This tissue too, contains both epithelium and stroma. The stroma contains many elements, including c-fms expressing tumor associated macrophages, which are active players in the breast tumor microenvironment (Lin et al. 2001). It has been shown that the “normal” tissue adjacent to the breast tumor can be an active tumor microenvironment which can portend more impact on prognosis than the breast epithelium itself (Huang et al. 2016). Lastly, detection of activated phosphorylated, c-fms protein, requires strict tissue collection processes. All these factors help explain the finding that c-fms protein expression detected in this way was down-regulated in breast cancer tissues compared to “normal” tissues. What is clear from this analysis, however, is that under the same conditions, HMGXB3 protein levels in breast cancer tissues were increased compared to the “normals” (Fig. 7). This analysis of in vivo breast tissues, showing that c-fms and HMGXB3 expression patterns are opposite, is in line with our in vitro findings regarding the existence and implications of c-fms and HMGXB3 convergent gene transcription in breast cancer cells.
In conclusion, we are the first to report that c-fms and HMGXB3 mRNA 3′ end polymorphism is derived from the extended transcription termination, with the intergenic region between c-fms and HMGXB3 genes of 162 bp. These c-fms and HMGXB3 mRNAs with the 3′ end extensions, have poly(A)+ tails, and represent 1−4% of total c-fms and HMGXB3 mRNAs.
The closely juxtaposed c-fms and HMGXB3 gene pair generates extended 3′ end transcripts, which form sense-antisense RNA pairs regulating each other′s expression.
We find that miR-324-5p, via targeting HMGXB3 3′ UTR mRNA, up-regulates c-fms expression; while HuR, via interaction and stabilization of c-fms mRNA, down-regulates HMGXB3 expression. These sense-antisense RNA pairs result in reduction of mRNA and protein expression largely on the basis of mRNA instability. Lastly, knock-in of a strong poly(A) signal abolishes bidirectional mRNA 3′ end extension, thereby reducing sense-antisense RNA pairing, up-regulating both c-fms and HMGXB3 mRNA and protein expression.
For future studies, it will be important to explore how c-fms and HMGXB3 sense-antisense RNA pair is processed to siRNA to induce gene silencing. In TGS, the RNA binding protein vigilin, which binds pyrimidine-rich sequence in c-fms mRNA 3′ UTR (Woo et al. 2011), may play a role as it has been reported to induce heterochromatin formation (Zhou et al. 2008).
MATERIALS AND METHODS
Cell culture
BT20 breast cancer cells were cultured in MEM supplemented with 10% fetal bovine serum.
Northern analysis
Total cellular RNA was extracted using TRIzol (Invitrogen). Ten micrograms total RNA was fractionated in 1% agarose-formaldehyde gel and blotted to Hybond-N nylon membrane (Amersham). 32P-UTP labeled c-fms and HMGXB3 strand-specific antisense RNA probes were generated as described in Supplemental Figure S1. Briefly, either c-fms (Woo et al. 2009) or HMGXB3 (in this work) DNA was PCR amplified and the 3′ end was tagged with T7 RNA polymerase promoter. In vitro transcription was performed with 32P-UTP and T7 RNA polymerase to synthesize the 32P-labeled strand-specific antisense RNA probe. After in vitro transcription, DNase I was treated to remove DNA template. Northern hybridization was done in 0.75 M NaCl, 10× Denhardt's (0.2% w/v Ficoll, 0.2% w/v Polyvinylpyrrolidone, 0.2% BSA), 50% formamide, and 32P-labeled antisense RNA probe (2 × 107 cpm/mL) at 42°C, overnight. The membrane was washed in 2× SSC (0.3 M NaCl, 30 mM sodium citrate) and 0.1% SDS at room temperature, and 0.1× SSC and 0.1% SDS at 65°C, 60 min. The membrane was exposed on a storage phosphor screen (GE Healthcare) up to 96 h before image capture by Typhoon FLA7000 Biomolecular Imager (GE).
A list of primers is presented in Supplemental Table S1.
mRNA 3′ end mapping
Poly(A)+-RNA from BT20 cells was purified using 5′-biotin-linked oligo-dT25 and Dynabeads MyOne Streptavidin C1 (Invitrogen). A total of 10 ng poly(A)+-RNA was reverse transcribed with RT primers by M-MuLV Reverse Transcriptase (NEB) and PCR amplified by Pfu Polymerase (Promega) as described in detail in Supplemental Figure S2. PCR products were purified by Monarch PCR & DNA Cleanup Kit (NEB) and sequenced to confirm 3′ end extension.
A list of RT and PCR primers is presented in Supplemental Table S1.
Quantification of mRNA with 3′ end extension
RNA quantification is described in detail in Supplemental Figure S2.
Briefly, 10 ng poly(A)+-RNA was reverse transcribed in a reaction tube with three sequence-specific RT primers; that is, RT primer 1 or 4 from c-fms or HMGXB3 mRNA coding region, RT primer 2 or 5 from c-fms or HMGXB3 mRNA 3′ end extension, and RT primer 3 from GAPDH mRNA. RT transcripts were real-time PCR amplified with sybrgreen for quantification as described in Supplemental Figure S2.
A list of primers is presented in Supplemental Table S1.
SV40-PA knock-in by CRISPR/Cas9
To construct the plasmid used in the SV40-PA knock-in (KI), DNA sequences for the left homology arm and right homology arm of targeted c-fms and HMGXB3 genes were cloned into the pDonor-D01 (GeneCopoeia). sgRNA-pCas-Guide-EF1a-GFP was generated (Origene).
The following two sgRNAs were used for knock-in that targeted the 5′ and 3′regions in IR.
sgRNA 1—AATTCCGTGCACATCGTATG
sgRNA 2—GGAATTTGCAGGTACTCATG
To obtain the c-fms-SV40-PA-HMGXB3-KI BT20 cell lines, 3 × 106 cells per well were seeded in a six-well plate with MEM supplemented with 10% fetal bovine serum at 37°C, 5% CO2. The following day, transfection was carried out with Fugene (Promega). One day later, puromycin (1 µg/mL) was added to the cells to increase the KI efficiency. Within ∼2 wk, positive single colonies were picked up and transferred into 24-well plates. Briefly, genomic DNA was isolated. A total of 100 ng genomic DNA was used as a template for a PCR, and the PCR products were analyzed by Sanger sequencing (Supplemental Fig. S3A).
RNA half-life
To determine c-fms and HMGXB3 mRNA half-life in BT20 breast cancer cells, actinomycin-D (Act-D) chase experiments were performed with 5 µg/mL of Act D (Sigma) added to inhibit new transcription. Cells were harvested at 0, 2, 4, 8, and 24 h after Act D treatment, and total cellular RNA was extracted using TRIzol (Invitrogen). qRT-PCR was performed.
c-fms and HMGXB3 mRNA half-lives were calculated after qRT-PCR, normalized to GAPDH mRNA, values were plotted, and the time period required for a given transcript to decrease to one-half of the initial abundance was calculated. GAPDH mRNA is not affected by miR-324-5p and has a long half-life (>18 h) (Woo et al. 2009). Three independent experiments were performed.
Gain-of-function and loss-of-function assays of miR-324-5p
Either hsa-miR-324-5p Mimic (Sigma HMI0479) or hsa-miR-324-5p Inhibitor (Sigma HSTUD0479) was transfected using MISSION siRNA Transfection Reagent (Sigma S1452) for 2 d. For negative control, MISSION miRNA, Negative Control (Sigma HMC0002) was transfected.
Luciferase reporter assay
Firefly luciferase Mut E in pcDNA3.1(−) was obtained from Robert J. Gillies (University of Arizona) (Baggett et al. 2004). Firefly luciferase Mut E fused with c-fms mRNA full length 3′ UTR (+3212 ∼ +3988) without 3′ end extension or HMGXB3 mRNA full length 3′ UTR (+4323 ∼ +5258) without 3′ end extension was constructed as described in Supplemental Figure S5.
For luciferase analysis, BT20 cells were transiently cotransfected using Fugene HD (Roche) with firefly luciferase-c-fms or HMGXB3 mRNA 3′ UTR plasmid and Renilla luciferase control plasmid (Promega). Dual luciferase-activity assays were performed 48 h after transfection according to the manufacturer's directions (Promega). Firefly luciferase activity was normalized by Renilla luciferase. Firefly and Renilla luciferase mRNAs, and GAPDH mRNA were measured by qRT-PCR. Firefly luciferase mRNA was normalized by Renilla luciferase mRNA, and followed by GAPDH mRNA.
RNA capture by immunoprecipitation
The BT20 cell was treated by formaldehyde for RNA–protein crosslink. Immunoprecipitation (IP) of the endogenous mRNP complex was done by the protocol described previously (Woo and Chambers 2019). For HuR IP, 5 µg mouse monoclonal anti-human HuR antibody (Santa Cruz, sc-5261) was used. A reaction containing 10 µg normal mouse IgG (Sigma) served as a negative control. For immunoblot, cytoplasmic protein was isolated using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific). Immunoblot was done with HuR antibody (Santa Cruz, sc-365816). For RNA isolation, IP material is digested by proteinase K for 30 min at 55°C, following phenol:CHCl3 extraction, and ethanol precipitation.
HuR overexpression and silencing in BT20 cells
HuR-pcDNA3.1 (Woo et al. 2009) was transfected for overexpression. For silencing, HuR shRNA (Origene, TI352933) was transfected using Fugene HD (Roche).
UALCAN analysis
UALCAN website (http://ualcan.path.uab.edu) is a comprehensive web resource for analyzing cancer OMICS data (Chandrashekar et al. 2017). We utilized this website to explore the protein expression of c-fms (i.e., CSF-1R) and HMGXB3 in breast cancer tissues, previously characterized by CPTAC in normal and malignant breast tissues from sources including the TCGA project.
Statistical analysis
Data are depicted as mean ± SD from at least three independent experiments. Exact n values are provided in the figure legends. The unpaired two-way t-test, nonlinear regression analysis, and one-way ANOVA were performed using SigmaStat (Jandel Scientific Corp.). P < 0.05 was considered statistically significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Women's Cancers of the University of Arizona Cancer Center and the Bobbi Olson Endowment Fund (to S.K.C.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078749.121.
REFERENCES
- Baggett B, Roy R, Momen S, Morgan S, Tist L, Morse D, Gillies RJ. 2004. Thermostability of firefly luciferase affects efficiency of detection by in vitro bioluminescence. Mol Imaging 3: 324–332. 10.1162/1535350042973553 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291. 10.1016/j.cell.2005.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D. 2007. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048. 10.1038/nsmb1315 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SK, Kacinski BM, Ivins CM, Carcangiu ML. 1997. Overexpression of epithelial CSF-1 and CSF-1 receptor: a poor prognostic factor in epithelial ovarian cancer; contrasted to a protective effect of stromal CSF-1. Clin Cancer Res 3: 999–1007. [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. 2017. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19: 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. 2004. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820. 10.1093/nar/gkh818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498. 10.1038/sj.emboj.7601932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E, ‘t Hoen PAC. 2015. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet 31: 128–139. 10.1016/j.tig.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. 10.1016/j.molcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378. 10.1016/s1097-2765(00)80464-5 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestead C. 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643. 10.1038/nrm2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick MB, Sapi E, Perrotta PL, Maher MG, Halaban R, Carter D, Kacinski BM. 1997. Recognition of activated CSF-1 receptor in breast carcinomas by a tyrosine 723 phosphospecific antibody. Oncogene 14: 2553–2561. 10.1038/sj.onc.1201092 [DOI] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995. 10.1016/j.cell.2008.02.040 [DOI] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. 2012. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol 19: 1193–1201. 10.1038/nsmb.2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Wang Y, Gao Y, Zhang Y, Chen M, Xu M, Hu L, Jing Y, Li C, Wang Q, et al. 2016. Knockdown of high mobility group-box 3 (HMGXB3) expression inhibits proliferation, reduces migration, and affects chemosensitivity in gastric cancer cells. Med Sci Monit 22: 3951–3960. 10.12659/msm.900880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans H, Alwine JC. 2000. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol Cell Biol 20: 2926–2932. 10.1128/mcb.20.8.2926-2932.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. 2012. RNA polymerase II collision interrupts convergent transcription. Mol Cell 48: 365–374. 10.1016/j.molcel.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Stern DF, Zhao H. 2016. Transcriptional profiles from paired normal samples offer complementary information on cancer patient survival – evidence from TCGA Pan-Cancer data. Sci Rep 6: 20567. 10.1038/srep20567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacinski BM, Carter D, Mittal K, Kohorn EI, Bloodgood RS, Donahue J, Donofrio L, Edwards R, Schwartz PE, Chambers JT, et al. 1988. High level expression of fms proto-oncogene mRNA is observed in clinically aggressive human endometrial adenocarcinomas. Int J Radiat Oncol Biol Phys 15: 823–829. 10.1016/0360-3016(88)90113-7 [DOI] [PubMed] [Google Scholar]
- Kacinski BM, Carter D, Mittal K, Yee LD, Scata KA, Donofrio L, Chambers SK, Wang KI, Yang-Feng T, Rohrschneider LR, et al. 1990. Ovarian adenocarcinomas express fms-complementary transcripts and fms antigen, often with coexpression of CSF-1. Am J Pathol 137: 135–147. [PMC free article] [PubMed] [Google Scholar]
- Kacinski BM, Scata KA, Carter D, Yee LD, Sapi E, King BL, Chambers SK, Jones MA, Pirro MH, Stanley ER, et al. 1991. FMS (CSF-1 receptor) and CSF-1 transcripts and protein are expressed by human breast carcinomas in vivo and in vitro. Oncogene 6: 941–952. [PubMed] [Google Scholar]
- Kainov YA, Aushev VN, Naumenko SA, Tchevkina EM, Bazykin GA. 2016. Complex selection on human polyadenylation signals revealed by polymorphism and divergence data. Genome Biol Evol 8: 1971–1979. 10.1093/gbe/evw137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Nam JW. 2006. Genomics of microRNA. Trends Genet 22: 165–173. 10.1016/j.tig.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C. 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 1–12. 10.1038/nrm3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. 2006. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO J 7: 1216–1222. 10.1038/sj.embor.7400857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Nguyen AV, Russell RG, Pollard JW. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med 193: 727–740. 10.1084/jem.193.6.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, McAllister WT. 2009. In a head-on collision, two RNA polymerases approaching one another on the same DNA may pass by one another. J Mol Biol 391: 808–812. 10.1016/j.jmb.2009.06.060 [DOI] [PubMed] [Google Scholar]
- Maher MG, Sapi E, Turner B, Gumbs A, Perrotta PL, Carter D, Kacinski BM, Haffty BG. 1998. Prognostic significance of colony-stimulating factor receptor expression in ipsilateral breast cancer recurrence. Clin Cancer Res 4: 1851–1856. [PubMed] [Google Scholar]
- Nourse J, Spada S, Danckwardt S. 2020. Emerging roles of RNA 3′-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules 10: 915. 10.3390/biom10060915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorodnikov A, Kargapolova Y, Danckwardt S. 2016. Processing and transcriptome expansion at the mRNA 3′ end in health and disease: finding the right end. Pflugers Arch Eur J Physiol 468: 993–1012. 10.1007/s00424-016-1828-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM. 2013. Gene regulation by antisense transcription. Nat Rev Genet 14: 880–893. 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci 14: 105–110. 10.1016/0968-0004(89)90132-1 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. 2009. Transcription termination by nuclear RNA polymerase. Genes Dev 23: 1247–1269. 10.1101/gad.1792809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapi E, Flick MB, Gilmore-Hebert M, Rodov S, Kacinski BM. 1995. Transcriptional regulation of the c-fms (CSF-1R) proto-oncogene in human breast carcinoma cells by glucocorticoids. Oncogene 10: 529–542. [PubMed] [Google Scholar]
- Schek N, Cooke C, Alwine JC. 1992. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol 12: 5386–5393. 10.1128/mcb.12.12.5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing M, Wang B, Feng G, Duan L, Yuan S, Jia W, Liu Y. 2019. Knockdown of high mobility group box 3 impairs cell viability and colony formation but increases apoptosis in A549 human non-small cell lung cancer cells. Oncol Lett 17: 2937–2945. 10.3892/ol.2019.9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Navickas A, Wery M, Descrimes M, Morillon A, Torchet C, Benard L. 2015. Cytoplasmic control of sense-antisense mRNA pairs. Cell Rep 12: 1853–1864. 10.1016/j.celrep.2015.08.016 [DOI] [PubMed] [Google Scholar]
- Song N, Wang B, Feng G, Duan L, Yuan S, Jia W, Liu Y. 2019. Knockdown of high mobility group box 3 impairs cell viability and colony formation but increases apoptosis in A549 human non-small cell lung cancer cells. Oncol Lett 17: 2937–2945. 10.3892/ol.2019.9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LN, Xing C, Zhi Z, Liu Y, Chen LY, Shen T, Zhou Q, Liu YH, Gan WJ, Wang JR, et al. 2017. Dicer suppresses cytoskeleton remodeling and tumorigenesis of colorectal epithelium by miR-324-5p mediated suppression of HMGXB3 and WASF-2. Oncotarget 8: 55776–55789. 10.18632/oncotarget.18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy EP, Chambers JT, Kacinski BM, Flick MB, Chambers SK. 2001. The activated macrophage colony-stimulating factor (CSF-1) receptor as a predictor of poor outcome in advanced epithelial ovarian carcinoma. Gynecol Oncol 80: 194–200. 10.1006/gyno.2000.6070 [DOI] [PubMed] [Google Scholar]
- Werner A, Sayer JA. 2009. Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens 18: 343–349. 10.1097/MNH.0b013e32832cb982 [DOI] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. 2008. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell 29: 600–610. 10.1016/j.molcel.2007.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HH, Chambers SK. 2019. Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech 1862: 35–46. 10.1016/j.bbagrm.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Woo HH, Zhou Y, Yi X, David CL, Zheng W, Gilmore-Hebert M, Kluger HM, Ulukus EC, Baker T, Stoffer JB, et al. 2009. Regulation of non-AU-rich element containing c-fms proto-oncogene expression by HuR in breast cancer. Oncogene 28: 1176–1186. 10.1038/onc.2008.469 [DOI] [PubMed] [Google Scholar]
- Woo HH, Yi X, Lamb T, Menzl I, Baker T, Shapiro DJ, Chambers SK. 2011. Posttranscriptional suppression of proto-oncogene c-fms expression by vigilin in breast cancer. Mol Cell Biol 231: 215–225. 10.1128/MCB.01031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol 21: 379–386. 10.1038/nbt808 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465–3475. 10.1093/nar/gkl473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang Q, Chen LL, Carmichael GG. 2008. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA 14: 1773–1781. 10.1261/rna.1036308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.