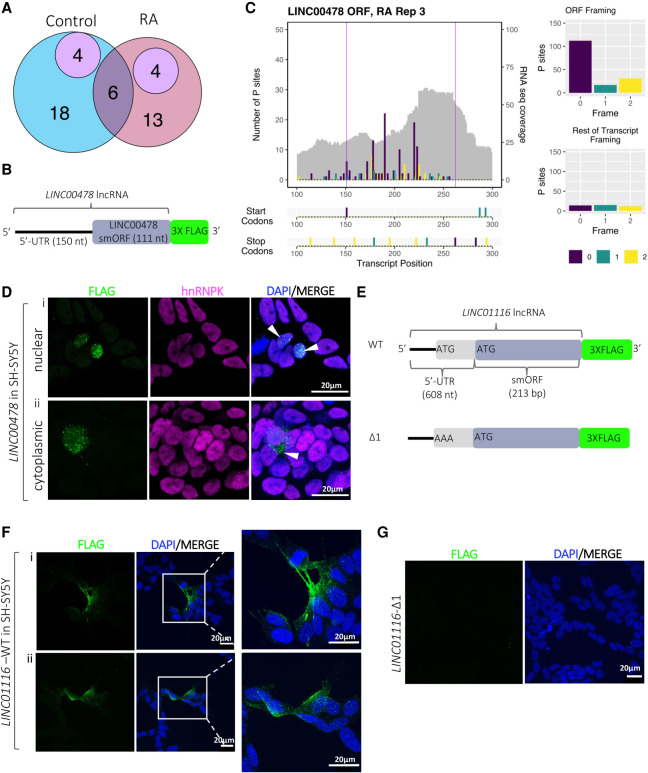
FIGURE 4.
Peptide production from smORFs in lncRNAs. (A) Venn diagram showing overlap lncRNA-smORFs detected between our Poly-Ribo-Seq and publicly available mass spectrometry data from SH-SY5Y (purple). Control in blue, RA in pink. (B) Schematic of tagging construct for LINC00478; lncRNA sequence upstream of smORF and smORF, excluding its stop codon, cloned upstream of 3× FLAG, which is lacking its own start codon. FLAG signal is therefore dependent on smORF translation. (C) Poly-Ribo-Seq profile for LINC00478 in RA treatment. RNA-seq (Polysome) reads are gray and ribosome P sites are in purple, turquoise, and yellow according to frame. Purple lines mark beginning and end of translated smORF. All possible start and stop codons are indicated below. Framing within and outside translated smORF shown on right. (D) Confocal images of FLAG-tagged LINC00478 peptide in SH-SY5Y cells (Control), showing (i) nuclear and (ii) cytoplasmic distribution, green is FLAG, magenta is hnRNPK (marking nuclei), and blue is DAPI (scale bar is 20 µm). (E) Schematic of tagging constructs for LINC01116 (WT and start codon mutant Δ1). (F) Confocal images of FLAG-tagged WT LINC01116 peptide showing cytoplasmic localization, near cell membrane and neuritic processes (magnification of insert is 3×). (G) Δ1 start codon mutant, showing no FLAG signal, in SH-SY5Y cells; green is FLAG and blue is DAPI (scale bar is 20 µm).