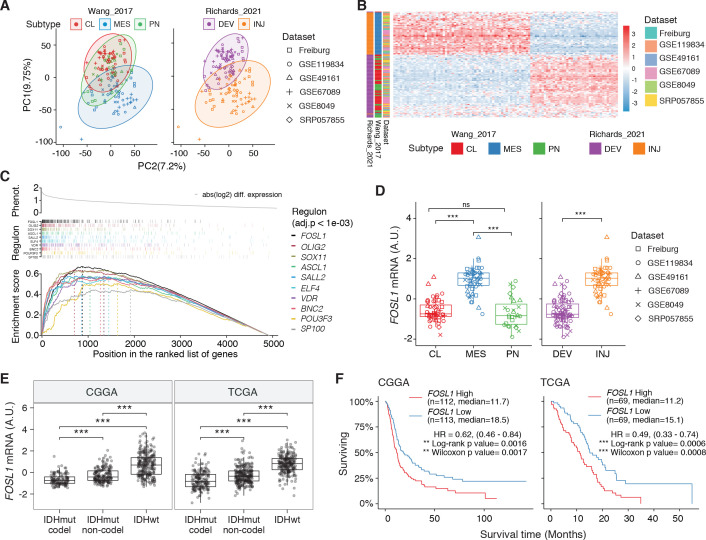
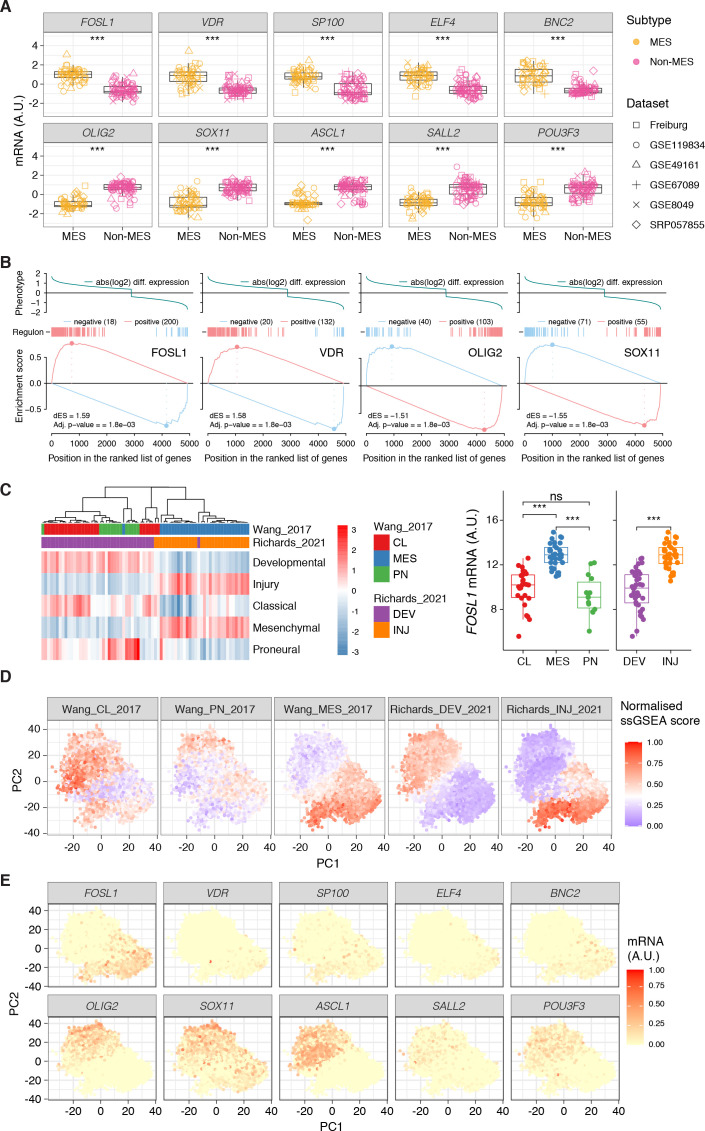
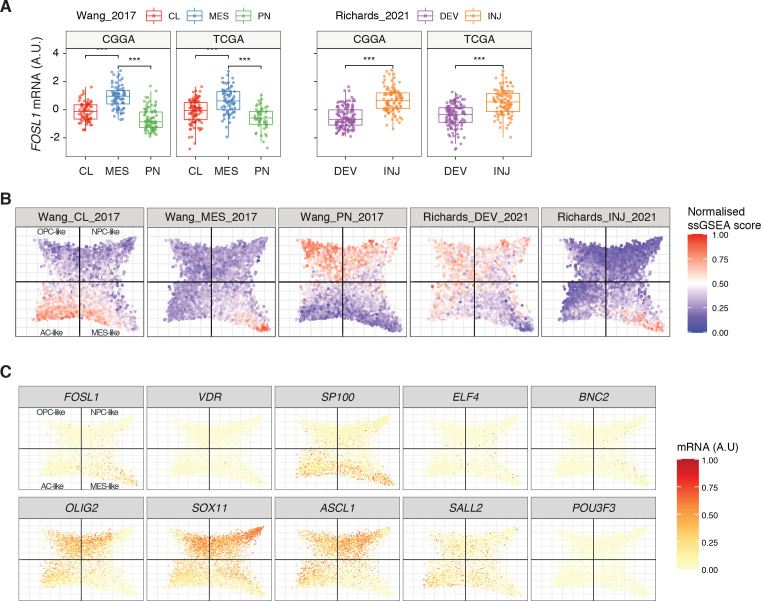
Figure 1. FOSL1 is a bona fide regulator of the glioma-intrinsic mesenchymal (MES) transcriptional signature.
(A) Principal component (PC) analysis of the brain tumor stem cells (BTSCs) expression dataset. (B) Heatmap of the top 100 differentially expressed genes between MES and non-MES BTSCs. (C) One-tail gene set enrichment analysis (GSEA) of the top 10 scoring transcription factors (TFs) in the master regulator analysis (MRA). (D) FOSL1 mRNA expression in the BTSCs dataset. One-way ANOVA with Tukey multiple pairwise comparison, ***p≤0.001, ns = not significant. (E) FOSL1 mRNA expression in the CGGA and TCGA datasets. Tumors were separated according to their molecular subtype classification. One-way ANOVA with Tukey multiple pairwise comparison, ***p≤0.001. (F) Kaplan–Meier survival curves of IDH-wt gliomas in the CGGA and TCGA datasets stratified based on FOSL1 expression (see Materials and methods for details).