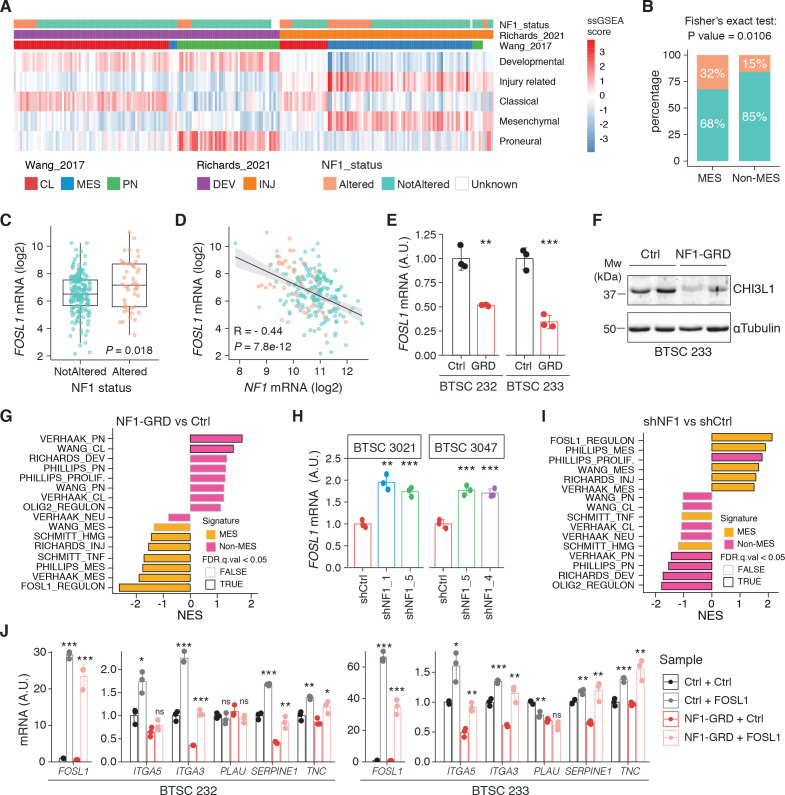
Figure 2. NF1 is a functional modulator of mesenchymal (MES) transcriptional signatures through FOSL1 expression regulation.
(A) Heatmap of the subtypes single-sample gene set enrichment analysis (ssGSEA) scores and NF1 genetic alterations of the IDH-wt gliomas in the TCGA dataset. (B) Frequency of NF1 alterations in MES and non-MES IDH-wt gliomas. Colors are as in panel (A). (C) FOSL1 mRNA expression in IDH-wt gliomas, stratified according to NF1 alterations. Colors are as in panel (A). Student’s t test, p=0.018. (D) Correlation of FOSL1 and NF1 mRNA expression in IDH-wt gliomas. Colors are as in panel (A). Pearson correlation, R = −0.044, p=7.8e-12. (E) qRT-PCR analysis of FOSL1 expression upon NF1-GRD overexpression in BTSC 232 and BTSC 233 cells. (F) Western blot analysis of whole-cell extract of BTSC 233 cells showing CHI3L1 mesenchymal marker expression upon NF1-GRD transduction; α-tubulin was used as loading control. Two biological replicates are shown. (G) Gene set enrichment analysis (GSEA) results of BTSC 233 cells transduced with NF1-GRD expressing lentivirus versus Ctrl. NES: normalized enrichment score. (H) qRT-PCR analysis of FOSL1 expression upon NF1 knockdown in BTSC 3021 and BTSC 3047 cells. (I) GSEA results of BTSC 3021 transduced with shNF1_5 versus Ctrl. (J) qRT-PCR analysis of MES genes expression upon NF1-GRD and FOSL1 co-expression in BTSC 232 and BTSC 233 cells. qRT-PCR data in (E), (H), and (J) are presented as mean ± SD (n = 3, technical replicates), normalized to 18S rRNA expression; Student’s t test, *p≤0.05, **p≤0.01, ***p≤0.001, ns = not significant.