Fig. 4. Interpreting the neural network.
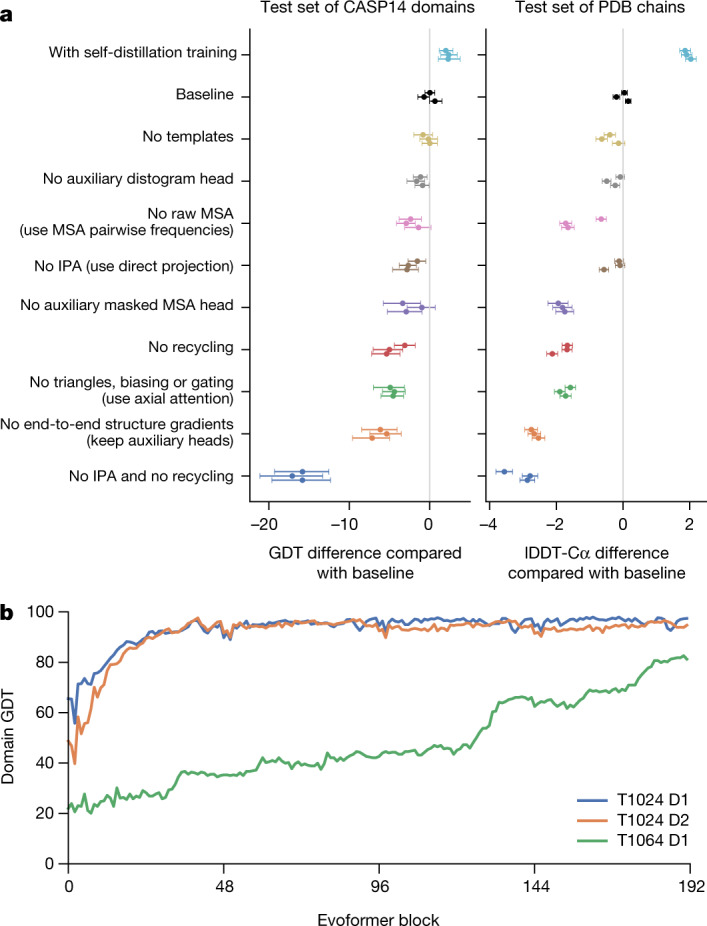
a, Ablation results on two target sets: the CASP14 set of domains (n = 87 protein domains) and the PDB test set of chains with template coverage of ≤30% at 30% identity (n = 2,261 protein chains). Domains are scored with GDT and chains are scored with lDDT-Cα. The ablations are reported as a difference compared with the average of the three baseline seeds. Means (points) and 95% bootstrap percentile intervals (error bars) are computed using bootstrap estimates of 10,000 samples. b, Domain GDT trajectory over 4 recycling iterations and 48 Evoformer blocks on CASP14 targets LmrP (T1024) and Orf8 (T1064) where D1 and D2 refer to the individual domains as defined by the CASP assessment. Both T1024 domains obtain the correct structure early in the network, whereas the structure of T1064 changes multiple times and requires nearly the full depth of the network to reach the final structure. Note, 48 Evoformer blocks comprise one recycling iteration.
