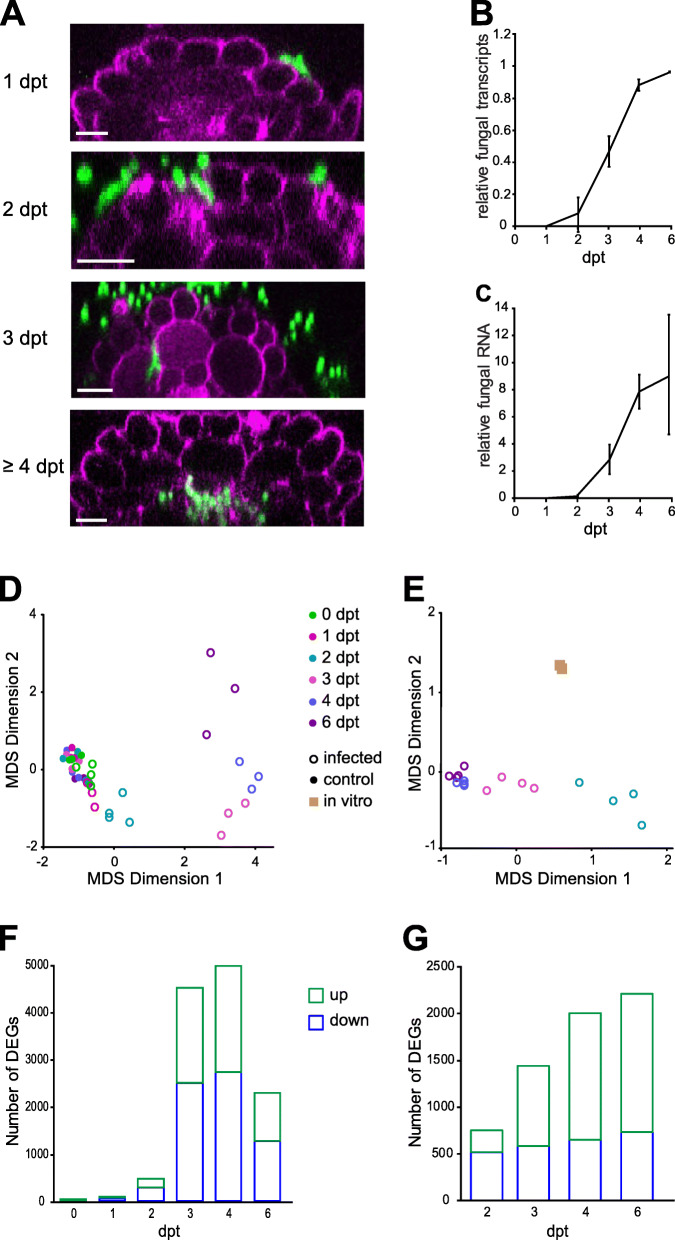
Fig. 1.
Fo5176 infection of Arabidopsis roots leads to temporal dynamic changes in the plant and the fungal transcriptomes. A Microscopic analysis of root infection by Fo5176 over 4 days after microconidia treatment (dpt). Left panels: Representative confocal images of Fo5176 (green) colonizing the different cell layers of Arabidopsis roots (magenta). Scale bars 20 μm. Each image represents a minimum of 9 hydroponically infected and 4 plate-infected roots/dpt. B Percentage of transcripts based on the RNA-seq analysis mapped to the fungal reference genome at all investigated time points using a splice aware sequence aligner (STAR). Values represent the mean ± standard error of four biological replicates. C Fo5176 biomass determination over time based on RNA and determined by qRT-PCR using a fungus-specific primer (Fo5176 β-Tub) relative to an Arabidopsis reference gene (At GAPDH). Values represent the mean ± standard error of four biological replicates. D, E Multidimensional scaling analysis (MDS) of transcriptional profiles of Arabidopsis (D) and Fo5176 (E). F, G Number of differentially expressed genes (DEGs) upregulated (green) or downregulated (blue) at each root colonization stage (0-6 dpt and 2-6 dpt, respectively) in Arabidopsis (F) and Fo5176 (G)