Abstract
The diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection requires the detection of viral RNA by reverse transcription–polymerase chain reaction (RT-qPCR) performed mainly using nasopharyngeal swabs. However, this procedure requires separate analysis per each individual, performed in advanced centralized laboratory facilities with specialized medical personnel. In this study, an alternative approach termed “solid waste-based surveillance (SWBS)” was explored, in order to investigate SARS-CoV-2 infection in small communities through the indirect sampling of saliva left on waste. Sampling was performed at 20 different sites in Italy during the second peak of COVID-19. Three swabs were positive for SARS-CoV-2 using a published RT-qPCR protocol targeting the non-structural protein 14 region, and the viral load ranged 4.8 × 103–4.0 × 106 genome copies/swab. Amino acid substitutions already reported in SARS-CoV-2 sequences circulating in Italy (A222V and P521S) were detected in two positive samples. These findings confirmed the effectiveness of SWBS for non-invasive and dynamic SARS-CoV-2 surveillance.
Keywords: RNA detection, Saliva, Swab, SARS-CoV-2, Solid waste based surveillance
Graphical abstract
1. Introduction
After more than one year since its first detection in the city of Wuhan, China (CDC, 2020), coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to spread worldwide. Together with its major impact on health, with approximately 182 million infected individuals and 4 million deaths reported as on 30 June 2021 (WHO, 2021), COVID-19 has exerted serious stresses on the healthcare systems of several countries and caused evident and relevant economic and social consequences. Eurostat (2020) reported an average gross domestic product in the 2020 for the EU27 of −6.2%, with peaks in some EU member lower than −11%. Some relevant social impacts of COVID-19 pandemic have been discussed, including negative consequences on the psychological well-being of most exposed groups (e.g., children, students, health workers) (Saladino et al., 2020). Developing feasible approaches for the early detection of SARS-CoV-2 infection is of the utmost importance to restrain its spread in the population and contain related effects on health and society.
Many approaches are already available to study the spatiotemporal characteristics of infectious diseases, such as sentinel surveillance, clinical-based surveillance, questionnaires or surveys, hospital admission data, and mortality and morbidity analyses (Mao et al., 2020). However, most of these approaches are highly dependent on the efficiency of national and local data collection systems. Moreover, they do not account for asymptomatic or paucisymptomatic cases.
A possible approach to resolve the aforementioned problems and support the monitoring of health conditions of communities is based on the analysis of the main urban waste products. More than half of the world's population now lives in urban areas, increasingly in highly populated cities. By 2050, it is projected that more than two-thirds of the global population will live in urban areas (UN, 2015), and 27 rapidly growing megacities currently consume approximately 9% of the world's electricity, generate 13% of its waste, and host 7% of the global population (Kennedy et al., 2015). For these reasons, monitoring public health in these areas will be of paramount importance for preventing any future pandemic diseases.
Modern cities are akin to organisms in that they consume resources to meet the needs of their populations and support economic activities, such as foods, materials, resources, and energy, returning metabolites (e.g., products, energy) and waste products including wastewater and gaseous and airborne particles. Among the different urban waste products, wastewater has been used to investigate some health and lifestyle aspects of given communities such as illicit drugs, pharmaceuticals, pesticides, nicotine, caffeine, alcohol (Choi et al., 2018; Daughton, 2001; Zuccato et al., 2005), as well as pathogens (Sinclair et al., 2008) The analysis of wastewater to identify presence of biologicals or chemicals for the purpose of monitoring public health is described as Wastewater-based epidemiology (WBE).
Recent studies showed that detecting the SARS-CoV-2 coronavirus in wastewater can be a low-cost solution for tracking COVID-19 outbreaks. The presence of SARS-CoV-2 in untreated wastewater was reported by several authors in different countries (Ahmed et al., 2020; Fongaro et al., 2020; Medema et al., 2020; La Rosa et al., 2020, La Rosa et al., 2021a).
Another important urban waste product of interest for potential SARS-CoV-2 monitoring could be waste that was in contact with human saliva such as disposable glasses and plastic bottles. It is well known that SARS-CoV-2 can persist on different materials such as plastic (Corpet, 2021), and it is recognized that the hands are efficient vehicles of virus transmission (Hirose et al., 2020). The literature on the municipal solid waste (MSW) sector in relation to SARS-CoV-2 is lacking, and no studies have focused on the presence of the virus on urban waste have been conducted.
Based on this evidence, the present study investigated the possible presence of SARS-CoV-2 on waste contacted by human saliva, such as plastic coffee cups, plastic glasses, beverage cans, and plastic bottles typically discharged in snack bars, canteens, restaurants, malls, offices, schools, and other business and organizations. The rationale for this study was bolstered by previous studies suggesting that saliva specimens are potential and reliable candidates for COVID-19 diagnosis because collection is minimally invasive and it can reliably be self-performed, minimizing the chance of exposing healthcare workers in comparison to nasopharyngeal swabs (Majam et al., 2021; Rodríguez Flores et al., 2021; To et al., 2020). The possibility of using MSW as a new means for monitoring and detecting the presence of infectious diseases in communities was also proposed and discussed.
2. Materials and methods
2.1. Solid waste sampling
A graphical representation of the experimental design is presented in Fig. 1 .
Fig. 1.

Graphical representation of the experimental design.
RT-qPCR, reverse transcription–quantitative polymerase chain reaction.
Based on the credited diffusion pathway of SARS-CoV-2 through droplets and saliva, the investigated waste included plastic coffee cups, plastic glasses, beverage cans, and plastic bottles. In particular, 20 different collection points were selected based on their generation of the different waste considered in this study. The collection points were located in a northern Italian Region significantly affected by the COVID-19 outbreak, and sampling was performed from January 9th, 2021 to February 20th, 2021 during the second pandemic peak. The geographic distribution of the sampling points encompassed a 570 km2 with approximately 630,000 inhabitants (~13% of the population of the Region) and, according to national statistics, population structure of the area under investigation (sex and age distribution) was in line with the regional ones (MID, 2021). During the sampling period an average of about 1050 new infected subjects per day (range 440–3100) was recorded in the Region with a steady decrease of cases up to late January, a stable incidence up to mid-February and a slight increase in correspondence of the end of the investigation (MID, 2021). Similarly, a reduction of the total number of the positivity rate (number of positive tests on the daily tests performed in the Region) from ~15% to ~2.5% between January 9th and February 20th was reported. With due caution and assuming even spread of SARS-CoV-2 infection within the Region, these figures can be expected to represent the infection trend in the study area.
At each collection point, a representative and standardized number of pieces ranging from 50 to 100 units were sampled and transported to the laboratory. Collected pieces that were excessively crumpled or contaminated by organic materials were excluded from the sampled pieces. Each piece was withdrawn from the bin using sterile gloves or clamps, inserted into double polyethylene bags, and transported under controlled temperature (4 °C) to the laboratory (maximum transport time: 2 h). Before samples were collected, liquid residues were carefully removed by tilting samples to one side to limit contamination of the whole edge.
2.2. Surface swab collection
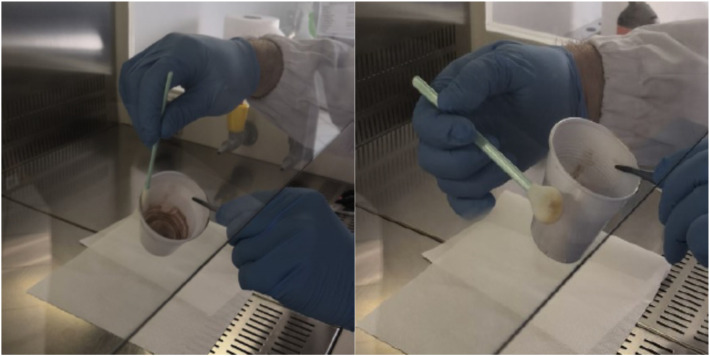
To focus sampling procedure on saliva residues from potentially infected subjects the tested objects were wiped only on the areas coming into contact with the users' mouths (e.g. cup and glass rims, bottle edges, etc.). Once delivered to the laboratory, swabs moistened with sterile saline solution were used to carefully wipe these areas; for the sampling operations, Texwipe Sterile CleanFoam Swabs (Texwipe, Kernersville, NC, USA) with a circular head of 9.4 mm in diameter (STX708A) were used, with each one employed to analyze a pooled sample of 50–100 pieces (Fig. 2 ). The 20 swabs were frozen at −80 °C for long-term storage and shipped under controlled time–temperature conditions (24 h, 4 °C) to Istituto Superiore di Sanità for molecular analyses, which were performed immediately upon arrival.
Fig. 2.
Swab testing procedure.
2.3. RNA extraction, real-time RT-qPCR, and nested RT-PCR
Nucleic acids were extracted from swabs using the NucliSens MiniMAG system (bioMérieux, Marcy-l'Étoile, France) according to the manufacturer's instructions with slight modifications (the use of 4 ml of lysis buffer for each swab and a lysis time of 20 min). Elution was performed in a 100-μl volume, and the extracted nucleic acids were further purified by PCR inhibitors using the OneStep PCR Inhibitor Removal Kit (Zymo Research, CA, USA). RNA was used immediately for molecular analysis and stored in aliquots at −80 °C.
Real-time RT-qPCR targeting the ORF1ab nsp14 region (3′-to-5′ exoribonuclease, ExoN) of the SARS-CoV-2 genome, previously developed for detection of the virus in wastewaters (La Rosa et al., 2021a) and validated using nasopharyngeal swabs (Pierri et al., 2021), was used to detect and quantify SARS-CoV-2 titers in the swabs. PCR inhibition was excluded by analyzing an external amplification control. Samples were also tested using the Logix Smart COVID-19 Test Kit (Co-Diagnostic, UT, USA) to confirm results with a second molecular target (SARS-CoV-2 RdRp gene) and assess the recovery of biological material from the surfaces of glasses and bottles/cans by analyzing human RNAse P.
Conventional RT-nested PCR amplification followed by sequencing was also performed for the molecular characterization of SARS-CoV-2 in the samples using an assay designed to detect key mutations of the spike protein (La Rosa et al., 2021b). The assay was a long-nested RT-PCR (approximately 1500 bps) protocol termed “PCR 979-980,” which was designed to screen for the presence of mutations characteristic of the three variants of concern (20I/501Y.V1, 20H/501Y.V2, and 20 J/501Y.V3), as well as other variants of interest. Briefly, reverse transcription for first-round PCR was performed in a 25-μl volume using 5 μl of RNA, 1 μl of each primer (10 μM), and the SuperScript III One-Step RT-PCR System with Platinum Taq (Invitrogen, CA, USA) with the following amplification conditions: 45 °C for 30 min; 94 °C for 2 min; 35 cycles of 94 °C for 15 s, 58 °C for 30 s, 68 °C for 1.75 min; and a final extension at 72 °C for 5 min. Nested PCR was performed in a 25-μl volume using Phusion Hot Start II DNA Polymerase with GC buffer (Thermo Fisher Scientific, MA, USA), 2 μl of the first PCR product, 1 μl of each primer (10 μM), and the following conditions: initial denaturation at 98 °C for 30 s; 45 cycles of 98 °C for 10 s, 62 °C for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 10 min. Molecular-grade water was included in each PCR run as a negative control. Good laboratory practice and standard precautions were taken to avoid contaminations.
PCR products were separated using 1% agarose gel electrophoresis and stained with ethidium bromide, and positive amplicons were purified from the PCR reaction or gel using the GRS PCR & Gel Band Purification Kit (GRISP, Porto, Portugal) and sequenced on both strands (Eurofins Genomics, Ebersberg, Germany). Sequences were submitted to GenBank under the accession numbers MZ209405-MZ209406.
3. Results
In total, 20 swab samples were tested for SARS-CoV-2. As each swab sample represented a pool variable from 50 to 100 units of waste coming in contact with human saliva, the tested swabs corresponded to a total of 1175 waste items and, roughly, to the same number of individual users. Three swabswere positive SARS-CoV-2 RNA according to real-time reverse transcription–quantitative polymerase chain reaction (RT-qPCR) targeting the non-structural protein 14 (nsp14) region of the SARS-CoV-2 genome (Table 1 ). The positive swab samples were taken from used plastic glasses and plastic coffee cups collected between February 9th, 2021 and February 19th, 2021 from snack bars positioned along main thoroughfares of the area included in the study. The number of pieces sampled collected in one pool each were 50, 75 and 50 for ID = 12, ID = 15 and ID = 19, respectively. The viral concentrations in the positive samples ranged 4.8 × 103–4.0 × 106 genome copies/swab, indicating the presence of considerable amounts of viral particles in human saliva. The presence of viral RNA in the three swabs was confirmed by commercial PCR targeting the RdRp gene of SARS-CoV-2. Concerning the reference gene ribonuclease (RNAse) P, the efficient recovery of human biological traces from the swabbed glasses and bottles was confirmed for 12/20 samples, with the cycle threshold (Ct) for positive results ranging from 28.11 to 38.99, and recovery was achieved both from plastic glasses/cups and bottles/cans. SARS-CoV-2–positive samples were associated with both high (Ct ≈ 28) and low (Ct ≈ 38) RNAse P recovery. Inhibition of PCR was ruled out by analyzing an external amplification control, and the values ranged 0%–20.3% (mean, 5.1%). Two of the three positive samples were successfully amplified and sequenced using a long-nested PCR targeting a portion of the spike protein. Compared to the Wuhan prototype strain, the two isolated strains featured the amino acid substitutions A222V and P521S.
Table 1.
Results of SARS-CoV-2 detection on waste and the molecular characterization of positive samples.
| Swab ID | Date | Waste type | SARS-CoV-2 |
Human RNAse P Ct | Nested RT-PCR ID 980 | Detected mutations (spike protein) | |
|---|---|---|---|---|---|---|---|
| nsp14 Ct (g.c./swab) | RdRp Ct | ||||||
| 1 | 09/01/2021 | Plastic coffee cups | – | – | 33.98 | – | |
| 2 | 12/01/2021 | Plastic coffee cups + plastic glasses | – | – | 35.27 | – | |
| 3 | 12/01/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 4 | 14/01/2021 | Plastic coffee cups + beverage cans | – | – | 37.40 | – | |
| 5 | 14/01/2021 | Plastic bottles + beverage cans | – | – | 33.99 | – | |
| 6 | 15/01/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 7 | 15/01/2021 | Plastic bottles + beverage cans | – | – | 34.71 | – | |
| 8 | 18/01/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 9 | 18/01/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 10 | 18/01/2021 | Plastic bottles + beverage cans | – | – | 33.74 | – | |
| 11 | 08/02/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 12 | 08/02/2021 | Plastic coffee cups | 22.68 (4.0E + 06) | 24.75 | 28.48 | + | A222V; P521S |
| 13 | 08/02/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 14 | 09/02/2021 | Plastic coffee cups | – | – | 35.51 | – | |
| 15 | 09/02/2021 | Plastic coffee cups + plastic glasses | 32.60 (4.8E + 03) | 34.20 | 37.91 | + | A222V; P521S |
| 16 | 12/02/2021 | Plastic bottles + beverage cans | – | – | – | – | |
| 17 | 12/02/2021 | Plastic coffee cups + plastic glasses | – | – | – | – | |
| 18 | 13/02/2021 | Plastic coffee cups + plastic glasses | – | – | 38.99 | – | |
| 19 | 19/02/2021 | Plastic coffee cups | 29.36 (4.3E + 04) | 32.05 | 28.11 | – | |
| 20 | 20/02/2021 | Plastic coffee cups | – | – | 36.89 | – | |
4. Discussion
In this study, an alternative approach to investigate the presence of SARS-CoV-2 in small communities was explored, through the indirect sampling of saliva on the inanimate surfaces of solid waste samples such as plastic coffee cups, plastic glasses, beverage cans, and plastic bottles. Furthermore, applying the pooled sampling of glasses and bottles, each sample (swab) represented a community of 50–100 individuals. With this sampling plan, 3 out of 20 swabs (i.e. 15%), corresponding to 175 waste items out 1175 sampled pieces, tested positive for SARS-CoV-2. All the positive swabs were related to samplings performed between February 9th, 2021 and February 19th, 2021 in snack bars along high traffic roads. It is worth mentioning that from the 9th to the 25th of January, according to the criteria for monitoring the health risk associated to the COVID-19 epidemic (D.M., 2020), the risk for the region under study was defined ‘moderate’ (ISS, 2021). This evaluation imposed restrictions to both commercial activities (including coffee shops and, restaurants) and population movements. Afterwards, with the re-assessment of the risk to ‘low’, an increase of mobility was registered (https://covid19.apple.com/mobility), with a corresponding increase of the probability of SARS-CoV-2 detection in waste items in contact with human saliva. Considering the adopted methodology and the level of investigation (i.e. pooled swabs, non-randomized sampling, etc.), it was not possible to establish a direct correlation between the results of this study and the number of infected subjects detected by the microbiological e epidemiological monitoring in the investigated area. Further studies are therefore required to explore the potential of solid waste-based surveillance. By the way, considering also that after February 19th the regional risk becomes again “moderate” (ISS, 2021), the proposed approach showed some ability, to be further investigated, for the detection of the virus in given places also in correlation with the regulation adopted for the management and for the prevention of the COVID-19 spreading.
Concerning the analyses, highly variable signals were obtained in terms of viral concentrations. This may be attributable to differences in either viral recovery from the glass/bottle surfaces or viral shedding among SARS-CoV-2–positive subjects. To our knowledge, this is the first study demonstrating the presence of the SARS-CoV-2 genome on waste being exposed to saliva. Interestingly, sequence analysis of PCR amplicons revealed the presence of two amino acid substitutions in two positive samples. The spike amino acid mutation A222V is common in SARS-CoV-2 sequences. As of May 18, 2021, this mutation had been detected 159,236 times (10.48% of all samples with spike sequences) in 101 countries. In Italy, 5217 of 28,753 SARS-CoV-2 sequences deposited in GISAID (18.1%) harbored this amino acid substitution. A222V is characteristic of clade GV, lineage B.1.177 (GISAID) or 20E.EU1 (Nexstrain), which emerged in the early summer of 2020 in Spain and subsequently spread to multiple locations in Europe, possibly introduced into other countries by summer tourists (Hodcroft et al., 2020). Instead, based on GISAID data, the amino acid mutation P521S has only been detected 119 times (0.01% of all samples with spike sequences) in 25 countries to date. The first strain with this amino acid mutation was collected in April 2020 (hCoV-19/Australia/NSW2627/2020). This mutation has been detected in five sequences found in the Italian regions of Apulia, Abruzzo, and Veneto, and four of these sequences carry the A222V substitution.
Several studies suggested the use of body fluids, including saliva, for the detection of SARS-CoV-2. Indeed, saliva collection is minimally invasive, and samples can be collected directly from the tested individuals without the need of trained personnel, resulting in fewer risks to healthcare personnel than swab-based methods. A number of studies evaluated SARS-CoV-2 detection in saliva, and most of them found greater or similar (≤10% difference) sensitivities for saliva compared to nasopharyngeal and other swab-based methods (Tan et al., 2021). Some countries have implemented saliva testing for SARS-CoV-2 detection, and pooled-sample testing of saliva has been applied for mass SARS-CoV-2 infection screening programs in schools and universities (Oba et al., 2021; Bi et al., 2021). Via low-cost frequent testing of students and faculty, pooled saliva analysis enabled schools to determine whether transmission had occurred and supported data-driven decisions.
This study proposes an approach that can similarly identify the occurrence of SARS-CoV-2 in small communities by detecting viral RNA in saliva on glasses, bottles, and cans. An advantage of this method is that it does not require individual sampling of saliva (and subsequent pooling) because pooling is performed using a single swab to test multiple units, creating a cost-efficient procedure. This method can therefore be used to rapidly monitor small communities such as offices, schools, canteens, malls, residential social and health facilities, and sports centers. A further advantage of this approach is that relies on indirect sampling, and therefore, it can be performed without additional gathering of the community under observation, thereby limiting exposure among sampling personnel and reducing the requirements for personal data protection, as samples are not associated with individual subjects (i.e., non-intrusive and privacy protection). Indeed, sampling of waste contaminated by saliva may be easily performed in areas and places after the end of operating hours, and based on the obtained results, individual screening within the community may be performed. More research is required to assess if such a community surveillance approach may be integrated with the use, in place of molecular tests, of antigenic rapid tests, which may be performed on site.
The presence of the SARS-CoV-2 genome in solid waste may raise questions regarding possible risks linked to the handling of these objects. It is known that contaminated fomites may play, even if in a limited capacity, a role in the spread of viruses in a wide range of environments, including hospitals, nursing homes, schools, offices, and restaurants (Goldman, 2020; Mondelli et al., 2021). The persistence of SARS-CoV-2 on several surfaces (e.g., steel, plastic, wood, and paper) ranges from a few hours up to several days during laboratory tests (Chin et al., 2020; Kampf et al., 2020; van Doremalen et al., 2020). Viral transmission can occur when a subject touches a contaminated fomite and then touches his/her mucosae. For a given item, the risk of contamination depends on different factors related to the interaction between viruses and materials, environmental conditions, the item's distance from an infected individual, and its frequency of handling. Solid waste such as plastic glasses and/or bottles is a disposable material that is discarded by individuals after use in appropriate trash containers. Therefore, based on the collection systems implemented in Italy as in the EU, these objects are unlikely to be further manually handled by other individuals. Furthermore, collection operators who handle the waste by road containers, bins and/or plastic bags are usually equipped with adequate individual protection devices, including gloves (Di Maria et al., 2020), and similar protection equipment are also adopted for the personnel operating in waste treatment facilities. As reported by some authors in relation with SARS-CoV-2 presence in wastewater (El Baz and Imziln, 2020; Gormley et al., 2020), some risk of contamination could also arise from the possible generation of aerosol during treatment of contaminated waste. By the way, there is no evidence of the possible role of this further transmission route for virus spreading.
In conclusion, this study proposes a strategy for the non-invasive monitoring of COVID-19 spread through the analysis of solid waste coming in contact with human saliva. The results support the use of this indirect sampling approach for the detection of SARS-CoV-2 infection in small communities and suggest additional research to assess the utility of this strategy for both small-scale and widespread monitoring plans.
5. Conclusions
The importance of efficient and effective surveillance methods for early detection of diseases has been widely demonstrated during this period of COVID-19 pandemic. Some approaches, as the one based on wastewater surveillance, are nowadays quite consolidated. Other approaches based on aerosols monitoring in internal as external environments have also been proposed but their feasibility is still to be demonstrated. Both these methods have been proposed for monitoring presence and trends of diseases in quite large communities. Otherwise, for the surveillance of narrowed communities as schools, canteens, factories and similar places, the analysis of some specific solid waste (i.e. plastic glasses, bottles and cans) that have been in contact with human saliva may be of particular interest. The results of this study confirmed that this last approach is technically feasible and that saliva specimens can be exploited for SARS-CoV-2 detection. Based on this evidence, solid waste-based surveillance (SWBS) appears as an approach worthy to be further investigated and implemented for the early detection of SARS-CoV-2 in small communities.
CRediT authorship contribution statement
Conception and design of study: Di Maria F, La Rosa G, Bonadonna L, Suffredini E, Pivato A.
Acquisition of data: La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Vincenza T, Suffredini E, Bonato T, Piazza R.
Analysis and/or interpretation of data: La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C., Iaconelli M, Beccaloni E, Scaini F, Bonadonna L, Vincenza T, Suffredini E, Bonato T.
Drafting the manuscript: Di Maria F, La Rosa G, Suffredini E, Bonadonna L, Bonato T, Pivato A, Piazza R.
Revising the manuscript critically for important intellectual content: Di Maria F, La Rosa G, Suffredini E, Bonadonna L, Pivato A.
Approval of the version of the manuscript to be published (the names of all authors must be listed): Di Maria F, La Rosa G, Bonato T, Pivato A, Piazza R, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Beccaloni E, Scaini S, Bonadonna L, Vincenza T, Suffredini E.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi C., Mendoza R., Cheng H.T., Pagapas G., Gabutan E., Khan N., Hoxie H., Holmes K., Gao N., Lewis R., Wang H., Neumann D., Chan A., Takizawa M., Lowe J., Chen X., Kelly B., Asif A., Barnes K., Khan N., May B., Wright M., Chowdhury T., Pollonini G., Gouda N., Guy C., Gordon C., Ayoluwa N., Colon E., Medzon M.N., Jones S., Hossain R., Dodson A., Wang M., McGaskey M., Vasileva A., Seibel S., Connolly J., Esposito M., Kim J., Lincoln A.E., Sikka R., Wyllie A.L., Berke E.M., Libien J., Pincus M., Premsrirut P.K. Pooled surveillance testing program for asymptomatic SARS-CoV-2 infections in K-12 Schools and Universities. medRxiv. 2021 doi: 10.1101/2021.02.09.21251464v1. published online Feb 12. https://www.medrxiv.org/content/ (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Centers of Disease Control and Prevention) 2020. Coronavirus Disease 2019 (COVID-2019)https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/summary.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fsummary.html Available at. [Google Scholar]
- Chin A.W.H., Chu J.T.S., Hui K.P.Y., Yen H.L., Chan M.C.W., Peiris M., Poon L.L.M., Perera M.R.A. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. Correspondance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. (Reference Ed.) 2018;105:453–469. [Google Scholar]
- Corpet D.E. Why does SARS-CoV-2 survive longer on plastic than on paper? Med. Hypotheses. 2021;146 doi: 10.1016/j.mehy.2020.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Illicit drugs in municipal sewage: proposed new non-intrusive tool to heighten public awareness of societal use of illicit/abused drugs and their potential for ecological consequences. Pharmaceuticals and personal care products on the environment: scientific and regulatory issue. Symposium. 2001;791:348–364. [Google Scholar]
- D.M. Adozione dei criteri relativi alle attivita' di monitoraggio del rischio sanitario di cui all'allegato 10 del decreto del Presidente del Consiglio dei ministri del 26 aprile 2020 – Introduction of the criteria concerning the monitoring activities of sanitary risk (In Italian) Off. J. Ital. Repub. G.U. Ser. Gen. 2020;(112) 02-05-2020. [Google Scholar]
- Di Maria F., Beccaloni E., Bonadonna L., Cini C., Confalonieri E., La Rosa G., Milana M.R., Testai E., Scaini F. Minimization of spreading of SARS-CoV-2 via household waste produced by subjects affected by COVID-19 or in quarantine. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baz S., Imziln B. Can aerosols and wastewater be considered as potentialtransmissional sources of COVID-19 to humans? Euro. J. Environ. Public Health. 2020;4(2) doi: 10.29333/ejeph/8324. [DOI] [Google Scholar]
- Eurostat GDP and main components (output, expenditure and income) 2020. https://ec.europa.eu/eurostat/databrowser/view/NAMA_10_GDP__custom_78848/bookmark/table?lang=en&bookmarkId=7681260e-2f75-4cd7-a153-02fc89543f2c Available at:
- Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P., Schorner M.A., Barazzetti F.H., Christoff A.P., de Oliveira L.F.V., Bazzo M.L. 2019. SARS-CoV-2 in human sewage in Santa Catalina. Sci. Total Environ. 2020;778 doi: 10.1016/j.scitotenv.2021.146198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet. 2020;20:892–893. doi: 10.1016/S1473-3099(20)30561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Correspondence. 2020;8 doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose R., Ikegaya H., Naito Y., Watanabe N., Yoshida T., Bandou R., Daidoji T., Itoh Y., Nakaya T. Survival of SARS-CoV-2 and influenza virus on the human skin: Importance of hand hygiene in COVID-19. Clin. Infect Dis. 2020 doi: 10.1093/cid/ciaa1517. Oct 3. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K., Bloom J.D., Veesler D., Vaughan T.G., Comas I., Candelas F.G., Stadler T., Neher R.A. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1101/2020.10.25.20219063. the preprint server for health sciences. [DOI] [Google Scholar]
- ISS Report monitoraggio settimanale - Weekly reports. 2021. https://www.iss.it/monitoraggio-settimanale Available at:
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C.A., et al. The energy and material flows of megacities. PNAS. 2015;112:5985–5990. doi: 10.1073/pnas.1504315112. http://www.pnas.org/content/112/19/5985.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brusaferro S., Brandtner D., Fasanella A., Pace L., Parisi A., Galante D., Suffredini E. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021;197:117104. doi: 10.1016/j.watres.2021.117104. Apr 2. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majam M., Msolomba V., Scott L., Stevens W., Marange F., Kahamba T., Venter F., Conserve D.F. Usability and clinical evaluation of self-sampling for SARS-CoV-2 diagnostic testing using nasal and saliva specimens in South Africa: study protocol. JMIR Res Protoc. 2021 doi: 10.2196/24811. Mar 9. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- MID Andamento nazionale COVID-19 – National trend of COVID-19. 2021. https://dati-covid.italia.it Available at:
- Mondelli M.U., Colaneri M., Seminari E.M., Baldanti F., Bruno R. Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. Lancet. 2021;21 doi: 10.1016/S1473-3099(20)30678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba J., Taniguchi H., Sato M., Takamatsu R., Morikawa S., Nakagawa T., Takaishi H., Saya H., Matsuo K., Nishihara H. RT-PCR screening tests for SARS-CoV-2 with saliva samples in asymptomatic people: strategy to maintain social and economic activities while reducing the risk of spreading the virus. Keio J. Med. 2021;2570(2):35–43. doi: 10.2302/kjm.2021-0003-OA. [DOI] [PubMed] [Google Scholar]
- Pierri B., Mancusi A., Proroga Y.T.R, Capuano F., Cerino P., Girardi S., Vassallo L., Lo Conte G., Tafuro M., Cuomo M.C., Di Concilio D., Vicenza T., Cozzi L., Di Pasquale S., La Rosa G., Beikpour F., Suffredini E. SARS-CoV-2 detection in nasopharyngeal swabs: performance characteristics of a real-time RT-qPCR and a droplet digital RT-PCR assay based on the exonuclease region (ORF1b, nsp 14) J. Virol. Methods. 2021 doi: 10.1016/j.jviromet.2021.114420. Submitted for pubblication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Flores S.N., Rodríguez-Martínez L.M., Reyes-Berrones B.L., Fernández-Santos N.A., Sierra-Moncada E.J., Rodríguez-Pérez M.A. Comparison between a standard and SalivaDirect RNA extraction protocol for molecular diagnosis of SARS-CoV-2 using nasopharyngeal swab and saliva clinical samples. Front. Bioeng. Biotechnol. 2021;29(9) doi: 10.3389/fbioe.2021.638902. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladino V., Algeri D., Auriemma V. The psychological and social impact of Covid-19: new perspectives of well-being. Front. Psychol. 2020;11 doi: 10.3389/fpsyg.2020.577684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249–269. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.H., Allicock O., Armstrong-Hough M., Wyllie A.L. Saliva as a gold-standard sample for SARS-CoV-2 detection. Lancet Respir. Med. 2021 doi: 10.1016/S2213-2600(21)00178-8. Published Online April 19, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M.-C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations . 2015. Adoption of the Paris Agreement. Framework Convention on Climate Change ‘Adoption of the Paris Agreement’. Paris, France. [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith M.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:12. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. COVID-19 Weekly Epidemiological Update. Available at Coronavirus disease (COVID-19) (who.int) (accessed 30.06.2021) [Google Scholar]
- Zuccato E., Chiabrando C., Castiglioni S., Calamari D., Bagnati R., Schiarea S., Fanelli R. Cocaine in surface waters: a new evidence-based tool to monitor community drug abuse. Environ. Health. 2005;5(4):14. doi: 10.1186/1476-069X-4-14. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]