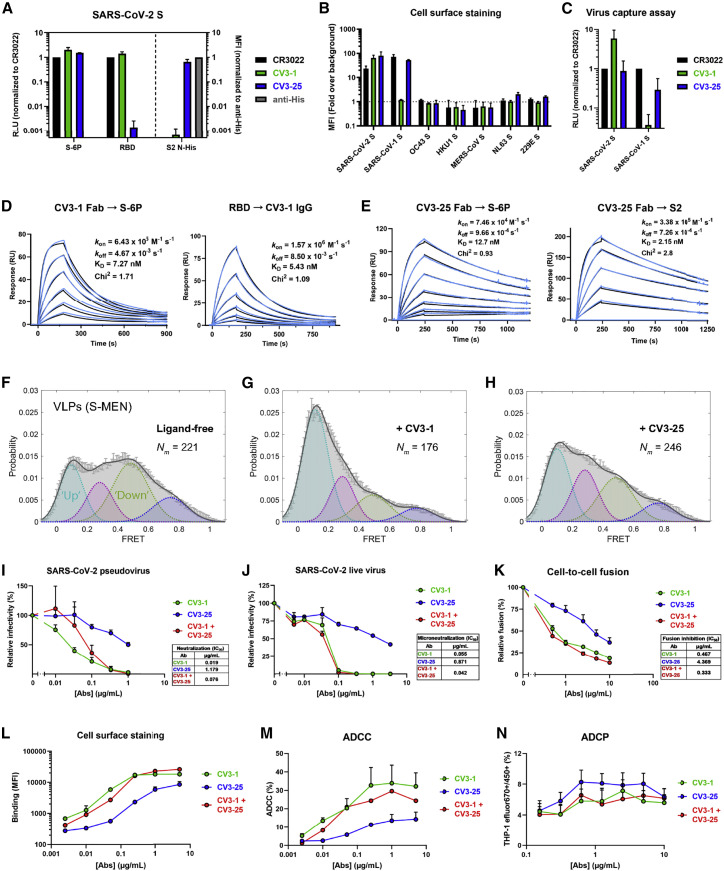
Figure 3.
CV3-1 and CV3-25 NAbs display potent neutralizing and antibody effector functions
(A) NAb binding to SARS-CoV-2 S ectodomain (S-6P) or RBD estimated by ELISA. Relative light units (RLU) were normalized to the cross-reactive SARS-CoV-1 mAb CR3022. NAb binding to SARS-CoV-2 S2 N-His tag protein on cell-surface of transfected 293T cells analyzed by flow cytometry. Median fluorescence intensities (MFIs) for anti-S NAbs were normalized to the signal obtained with an anti-His tag mAb.
(B) Flow cytometric detection of 293T cells expressing S from the indicated human CoVs. MFI from 293T cells transfected with empty vector was used for normalization.
(C) Pseudoviruses bearing SARS-CoV-2 or SARS-CoV-1 S were tested for capture by anti-S NAbs. The cross-reactive CR3022 mAb was used for normalization.
(D and E) NAb binding affinity and kinetics to SARS-CoV-2 S using surface plasmon resonance (SPR). SARS-CoV-2 S-6P or S2 ectodomain was immobilized as the ligand on the chip and CV3-1 (D) or CV3-25 (E) Fab was used as analyte at concentrations ranging from 3.12 to 100 nM and 1.56 to 100 nM, respectively, to S-6P and 3.125nM to 200nM for CV3-25 to S2 (2-fold serial dilution; see STAR Methods for details). Alternatively, CV3-1 immunoglobulin G (IgG) was immobilized on the chip and SARS-CoV-2 RBD used as analyte from 1.56 to 50 nM (2-fold serial dilution). Kinetic constants were determined using a 1:1 Langmuir model in bimolecular interaction analysis (BIA) evaluation software (experimental readings depicted in blue and fitted curves in black). Data shown are from one experiment.
(F–H) FRET histograms of ligand-free S on S-MEN coronavirus-like particles (VLPs) (F) or in presence of 50 μg/mL of CV3-1 (G) or CV3-25 (H). VLPs were incubated for 1 h at 37°C before smFRET imaging. Nm is the number of individual FRET traces compiled into a conformation-population FRET histogram (gray lines) and fitted into a 4-state Gaussian distribution (solid black) centered at 0.1-FRET (dashed cyan), 0.3-FRET (dashed red), 0.5-FRET (dashed green), and 0.8-FRET (dashed magenta). FRET histograms represent mean ± SEM (standard error). SEM is determined from three randomly distributed populations of all FRET traces under each indicated experimental condition.
(I) Neutralizing activity of CV3-1 and CV3-25 alone or in combination (1:1 ratio) on SARS-CoV-2 S bearing pseudoviruses using 293T-ACE2 cells.
(J) Microneutralization activity of anti-S NAbs on live SARS-CoV-2 virus using Vero E6 cells.
(K) Inhibition of cell-to-cell fusion between 293T cells expressing HIV-1 Tat and SARS-CoV-2 S and TZM-bl-ACE2 cells by NAbs.
Half maximal inhibitory antibody concentration (IC50) values in (I–K) were determined by normalized non-linear regression analyses.
(L) MFI of CEM.NKr cells expressing SARS-CoV-2 S (CEM.NKr-S) stained with indicated amounts of NAbs and normalized to parental CEM.NKr.
(M) Percentage of ADCC in the presence of titrated amounts of NAbs using 1:1 ratio of parental CEM.NKr cells and CEM.NKr-S cells as targets when PBMCs from non-infected donors were used as effector cells
(N) Percentage of ADCP in the presence of titrated amounts of NAbs using CEM.NKr-S cells as targets and THP-1 cells as phagocytic cells.
Results shown in panels (A–C), (I–K), and (N) were obtained in at least three or (L and M) two independent experiments