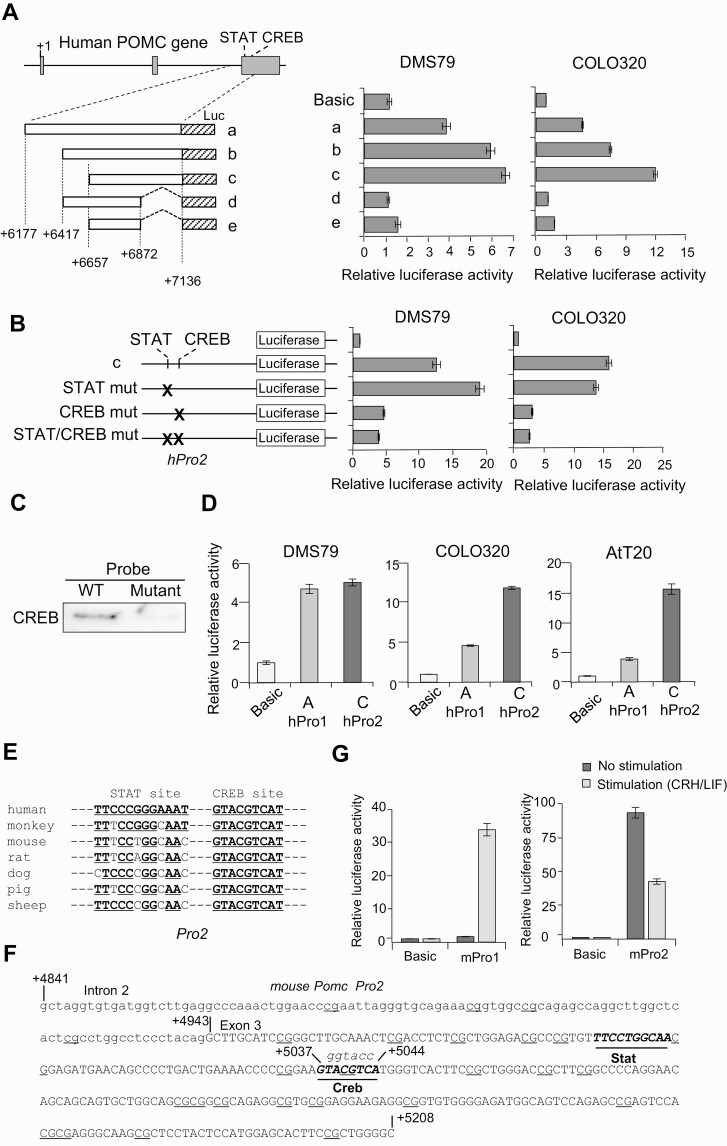
Figure 2.
Activity of the second proopiomelanocortin (POMC) promoter. A, Left, structure of 5 luciferase (Luc) reporter plasmids (a-e). The inserted POMC gene fragments are depicted as boxes, and positions of the 5′ and 3′ ends are noted. Right, luciferase assays were performed using indicated cells (DMS79 and COLO320) and generated luciferase activities with indicated plasmids compared with negative control plasmid (no promoter; Basic). B, Left, structure of luciferase reporter plasmids shown in A with mutated (mut) potential STAT, CREB, and STAT/CREB binding sequences (indicated by X) in fragment c in the second promoter (hPro2). Right, generated luciferase activities with indicated plasmids compared with negative control plasmid (no promoter; Basic) in DMS79 and COLO320 cells. C, Biotinylated probes from DNA binding assay with wild-type (WT) and mutated (mutant) CREB binding sites coprecipitated with proteins and analyzed by immunoblotting using anti-CREB. D, Luciferase assays using reporter plasmids containing the first promoter (hPro1) (–428 to +68) and second promoter (hPro2) (+6657 to +7136) compared with negative control plasmid (no promoter; Basic) using DMS79, COLO320, and AtT20 cell lines. E, Alignment of POMC second promoter STAT and CREB response elements by species. F, DNA sequence in mouse Pomc second promoter with the intron 2 sequence shown in lowercase letters and exon 3 sequence in capital letters. Potential STAT and CREB binding sequences are shown in italics and underlined, and dinucleotide 5′-CG-3′ (CpG) sequences identified as potential methylation sites are underlined. G, Luciferase assays using reporter plasmids mouse first promoter (mPro1) (–459 to +80) and mouse second promoter (mPro2) (+4841 to +5209) compared with negative control plasmid (no promoter; Basic) with CRH and LIF stimulation or without stimulation in AtT20 cells.