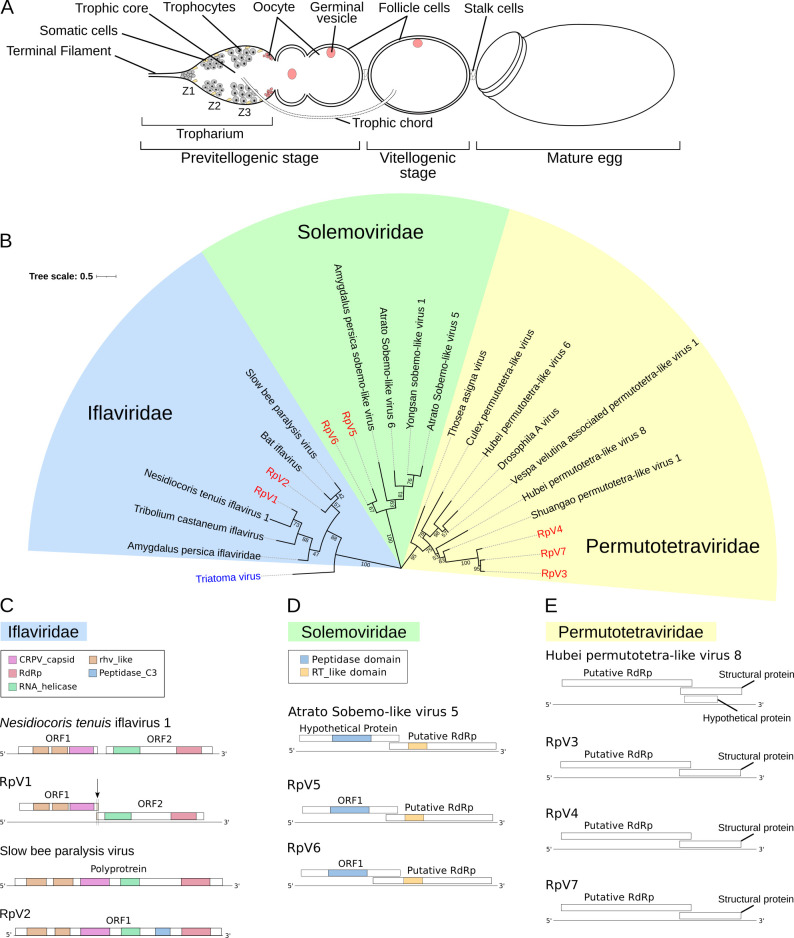
Fig 1. Phylogenetic analysis of the R. prolixus viruses.
(A) Schematic of an ovariole in R. prolixus. Mitotically active trophocytes (i.e. nurse cells) are harbored in Zone 1 of the tropharium, while polyploid trophocytes are present in Zone 2 and 3. The egg chamber in meroistic telotrophic ovaries is formed by the oocyte surrounded by somatic epithelial cells. Trophic cords connect the tropharium to the growing egg chamber and provide transport routes for nutrients and other molecules. (B) Phylogenetic tree of the RpVs constructed with RdRp sequences using Neighbor-Joining method with 1000 bootstrap replicates. Bootstrap values are displayed in tree branches. The RpVs are grouped in three families: Iflaviridae, Permutotetraviridae and Solemoviridae. (C) Genome organization of the RpV1 (MZ328304) and RpV2 (MZ328305) iflaviruses compared to the closest relatives Nesidiocoris tenuis iflavirus 1 and Slow bee paralysis virus respectively. RpV1 displays the 1nt frameshift typical of iflaviruses, that was not detected in the N. tenuis’ virus. (D) Diagram of the RpV3 (MZ328306), RpV4 (MZ328307) and RpV7 (MZ328310) and their closest relative Hubei permutotetra-like virus 8. (E) Genome organization of RpV5 (MZ328308), RpV6 (MZ328309) and Atrato Sobemo-like virus 5. rhv-like = Picornavirus/Calicivirus coat protein, CRPV_capsid = Cricket Paralysis Virus capsid protein, RdRp = RNA-dependent RNA Polymerase, RT_like = Reverse Transcriptase_like.