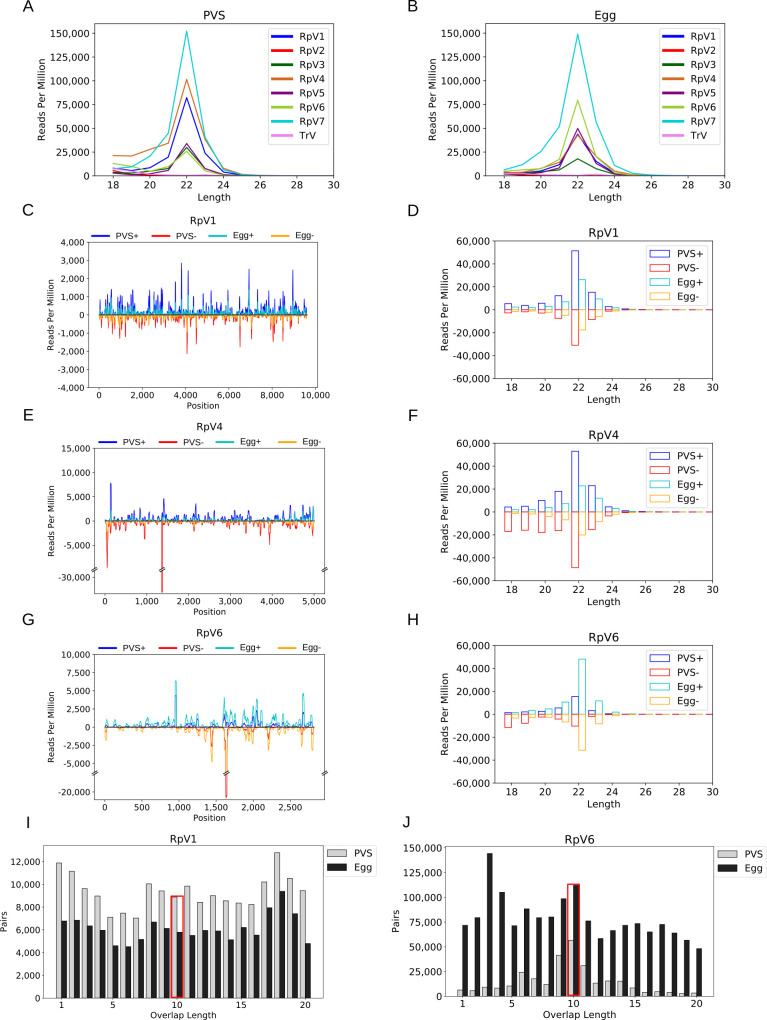
Fig 3. vsiRNA profiling during R. prolixus oogenesis.
(A) Length distribution of the RpVs small RNAs in previtellogenic stages (PVS) of oogenesis. (B) Length distribution of the viral small RNAs in mature eggs (Egg). In both stages of R. prolixus oogenesis, a peak at 22nt is readily detectable for all the RpVs, except for RpV2 and TrV, which did not generate vsiRNA reads above background levels. RpV2 was lost from our insectarium by the time that the small RNA datasets were produced. TrV was never detected in our colony and was used as control for the background small RNA levels. (C-H) vsiRNA profiling along the RpV1, RpV4 and RpV6 genomes and length distribution of the sense and antisense vsiRNAs. Sense and antisense vsiRNAs detected in previtellogenic stages of oogenesis (dark blue = sense, red = antisense) and mature chorionated eggs (light blue = sense, orange = antisense) are displayed for each virus. (I-J) Ping-pong signal for the RpV1 and RpV6 viruses in PVS (white bars) and Egg (grey bars). The red box highlights the position of the typical 10nt overlap between sense and antisense small RNA pairs.