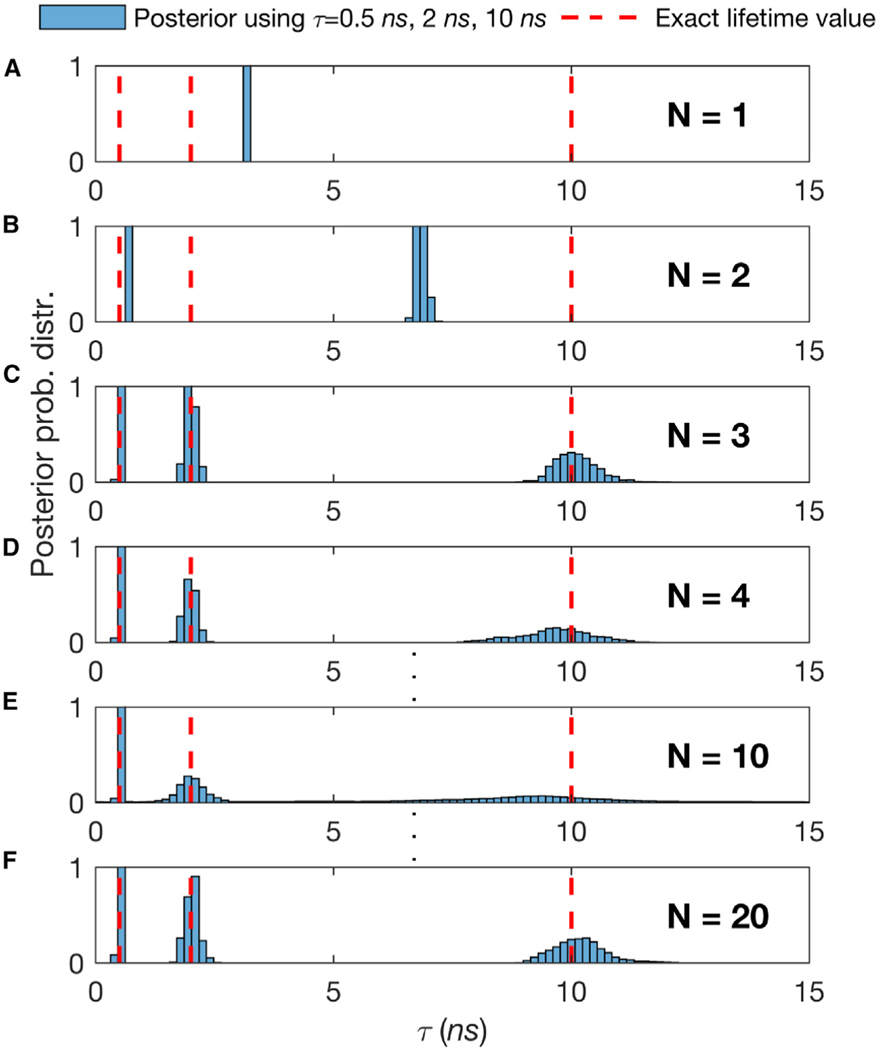
Figure 2. The Number of Species Assumed in the Analysis Directly Affects the Lifetimes Ascribed to Those Species, so an Independent Method Is Required to Estimate Species Numbers.
(A–F) We generate synthetic traces with 3 species with a total of 2 × 104 photon arrivals and lifetimes, τ, of 0.5, 2, and 10 ns. To estimate the τ within the normal (i.e., parametric) Bayesian paradigm, we start by assuming the following number of species, N= 1 (A), N= 2 (B), N= 3 (C), N= 4 (D), … , N= 10 (E), … , and N= 20 (F). The good fit provided by N > 2 and the mismatch in the peak of the posterior distribution over the lifetime and correct value of the lifetime (red dotted line) in all others underscores why it will become critical for us, or any method analyzing single photon data in the context of confocal microscope experiments, to correctly estimate the number of species contributing to the trace to deduce chemical parameters such as lifetime.