Figure 5.
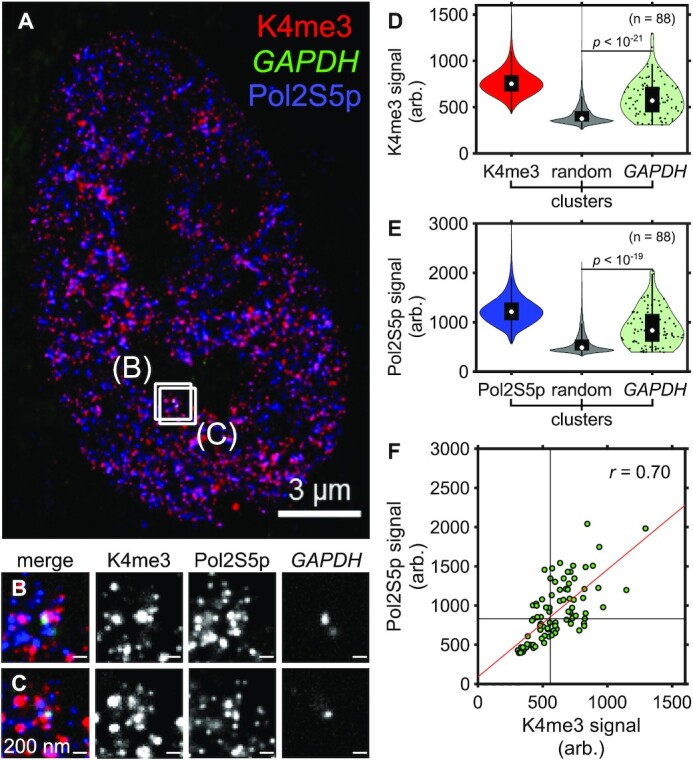
SCEPTRE compares H3K4me3 and paused RNA polymerase II signals at a single genomic region. (A) An expanded RPE1 cell with immunolabeled H3K4me3 marks (K4me3, red) and paused RNA polymerase II (Pol2S5p, blue), and FISH-labeled GAPDH (green). (B, C) Zoomed in views of the approximate center plane of an image stack for each GAPDH allele in the cell seen in (A). (D) Distributions of H3K4me3 fluorescence signal (arb. = arbitrary units) within H3K4me3, randomly selected regions (random) and GAPDH clusters. (E) Distribution of paused RNA polymerase II fluorescence signal within paused RNA polymerase II, randomly selected regions and GAPDH clusters. (F) H3K4me3 and paused RNA polymerase II fluorescence signals within GAPDH clusters (green). Black lines represent the threshold ‘on’ level for each fluorescence signal, while the red line represents the linear regression. The correlation coefficient (r) between fluorescence signals within GAPDH is shown in the top-right corner of the plot. Cluster numbers for (D). and (E). are K4me3 = 440298, Pol2S5p = 542245, random = 8240, GAPDH = 88. Significance determined by a right-tailed Wilcoxon rank-sum test of histone mark fluorescence signals in GAPDH against random cluster distributions. All scale bars are in pre-expansion units.
