Figure 7.
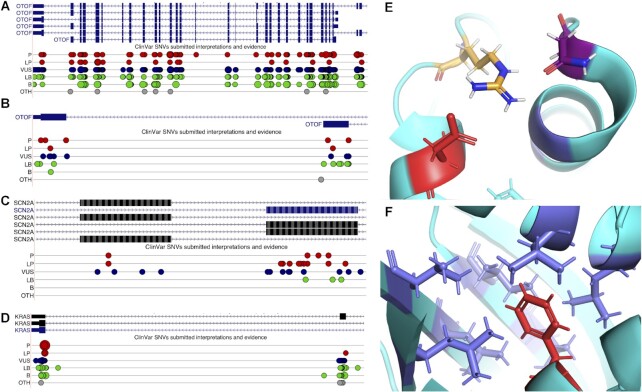
Tandem duplicated exons and ClinVar pathogenic mutations. (A) View of the gene OTOF from the UCSC Genome Browser, showing coding exons (wide blue rectangles), non-coding exons (narrower blue rectangles) and introns (blue lines with arrows). Mapped ClinVar mutations are shown below the transcripts; pathogenic (P) and likely pathogenic (LP) are shown as red spots, variants of unknown significance (VUS) are dark blue, likely benign (LB) and benign (B) as green and others (OTH) as grey. The larger the spots, the larger the mutation; most mutations are single nucleotide variations. (B) Close up view of two of the transcripts from OTOF, showing just the homologous 3′ coding exons; pathogenic and likely pathogenic mutations map to both exons. (C) Close up view of SCN2A transcripts in the UCSC Genome Browser, showing homologous mutually exclusively spliced coding exons; pathogenic and likely pathogenic mutations map to both exons. (D) Close up view of KRAS transcripts in the UCSC Genome Browser, showing homologous 3′ coding exons; pathogenic and likely pathogenic mutations map to one exon, a likely pathogenic mutation maps to the other. (E) Close up of one of the residues affected by pathogenic mutations in splice isoform KRAS4B (PDB: 6ms9). The mutation changes aspartate residue 153 (shown in red) to a glycine. The aspartate plays an important role in the structure of KRAS4B, forming a salt bridge with arginine 149 (orange and blue sticks), which in turns forms hydrogen bonds with the asparagine at residue 26 (purple and red sticks). The equivalent residue to aspartate 153 in isoform KRAS4A is a glutamate, and glutamate also forms salt bridges with arginine. The likely pathogenic mutation in KRAS4A, shown in section D, changes the glutamate to valine. Neither valine nor glycine form salt bridges. (F) Close up of the second residue affected by the pathogenic mutation in splice isoform KRAS4B (PDB: 6ms9). The mutation changes phenylalanine residue 156 (shown in red) to a valine (a much smaller hydrophobic amino acid). The phenylalanine nestles in a highly hydrophobic pocket (hydrophobic residues shown here as blue sticks), which is crucial for maintaining the structure (and therefore the function) of KRAS4B. Both KRAS4A and KRAS4B have a phenylalanine residue in this position.