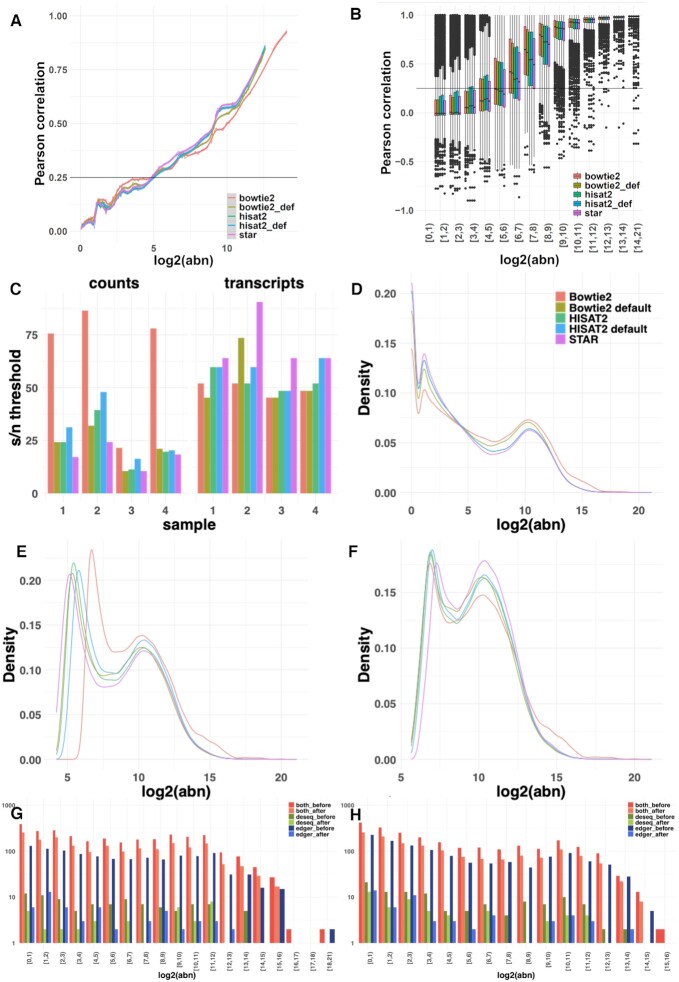
Figure 6.
Assessment of aligner choice on noise quantification. (A) The distribution of PCC across abundance bins in datasets for a single mRNAseq sample obtained by STAR, Bowtie2 and HISAT2 alignment followed by featureCounts quantification using a counts-based noise removal approach. (B) The distribution of PCC across abundance bins in aligned read counts obtained by the five aligners for the same sample in the transcript-based noise correction approach. (C) The detected signal-to-noise thresholds in the four mRNAseq samples varied when the counts or transcripts-based noise correction methods were applied. (D) The distribution of abundance of reads aligned by the five algorithms and quantified by featureCounts. (E) The distribution of abundance of the quantified counts after counts-based noise correction (F) The distribution of abundance of the quantified counts after transcripts-based noise correction. (G) The number of the differentially expressed genes found by applying DESeq and edgeR on the original and denoised (using transcripts-based approach) count matrices obtained by Bowtie2 alignment. (H) The overlap between the DESeq and edgeR analyses performed on the original and denoised counts matrices obtained by HISAT2.