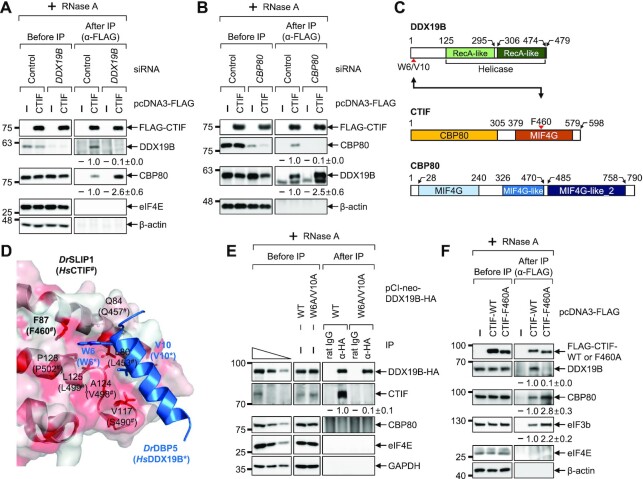
Figure 2.
CBP80 competes with DDX19B for binding to CTIF. (A) IP of FLAG-CTIF in the extracts of cells depleted of DDX19B. HEK293T cells either depleted or not depleted of endogenous DDX19B were transiently transfected with a plasmid expressing either FLAG or FLAG-CTIF-WT. The relative levels of coimmunoprecipitated DDX19B and CBP80 were quantified and are presented at the bottom of each image. Data are presented as the mean ± standard deviation; n = 2. (B) IPs of FLAG-CTIF in the extracts of cells depleted of CBP80. As performed in panel A, except that HEK293T cells were either depleted or not depleted of endogenous CBP80; n = 2. (C) Schematic illustration of DDX19B, CTIF and CBP80. CTIF consists of a CBP80-interacting domain (yellow) and a middle domain of eIF4G (MIF4G) domain (orange). The point mutations characterized in this study are indicated by red arrowheads. (D) Structure of a Danio rerio SLIP1 (DrSLIP1) and DrDBP5 complex (PDB: 4JHJ). The hydrophobic property is presented in the molecular surface of DrSLIP1: the more intense the red color is, the stronger is the hydrophobicity. Ribbon diagrams of DrDBP5 are shown in blue. Two important residues (W6 and V10) of DrDBP5 for interacting with DrSLIP1 are specified. In addition, the amino acid residues involved in the hydrophobic interaction between DrDBP5 and DrSLIP1 are indicated. The predicted amino acid residues important for an interaction between Homo sapiens CTIF (HsCTIF; homologous to DrSLIP1) and HsDDX19B (homologous to DrDBP5) are indicated in brackets. (E) IP of DDX19B-WT-HA or its W6A/V10A variant. Extracts of HEK293T cells expressing either DDX19B-WT-HA or DDX19B-W6A/V10A-HA were subjected to IPs with either α-HA antibody or rat IgG; n = 2. (F) IP of FLAG-CTIF or its F460A variant. Extracts of HEK293T cells expressing FLAG, FLAG-CTIF-WT or FLAG-CTIF-F460A were subjected to IP with the α-FLAG antibody; n = 2.