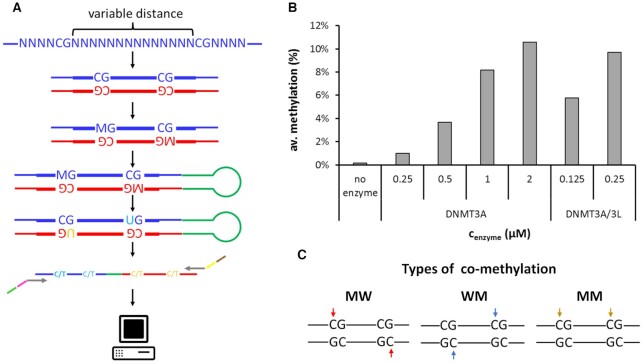
Figure 1.
DNMT3A and DNMT3A/3L catalyzed co-methylation of CpG sites in different distances. (A) Schematic view of the Deep-Enzymology workflow applied here. A library of synthetic oligonucleotides with CpG sites in variable distances embedded in random flanking sequence context is synthesized. After second strand synthesis, the pool of substrates with different sequences is methylated by DNMT3A or DNMT3A/3L, followed by hairpin ligation and bisulfite conversion. Barcodes and indices are added by PCR and the library is subjected to Illumina NGS sequencing. After quality control of the obtained reads, the original sequence and methylation state is extracted followed by analysis with respect to co-methylation events and sequence enrichment. M represents 5mdC. Methylation in the upper DNA strand is indicated by ‘M’, methylation in the lower DNA strand by ‘W’. (B) Overall methylation levels in the DNA methylation experiments with different enzyme concentrations and in the no-enzyme control. The data show average methylation levels at all 4 target cytosines in the two CpG sites averaged for all distances. (C) Schematic illustration of the different types of co-methylation of pairs of CpG sites considered in this study. Note that mechanistically MM and WW methylation are equivalent.