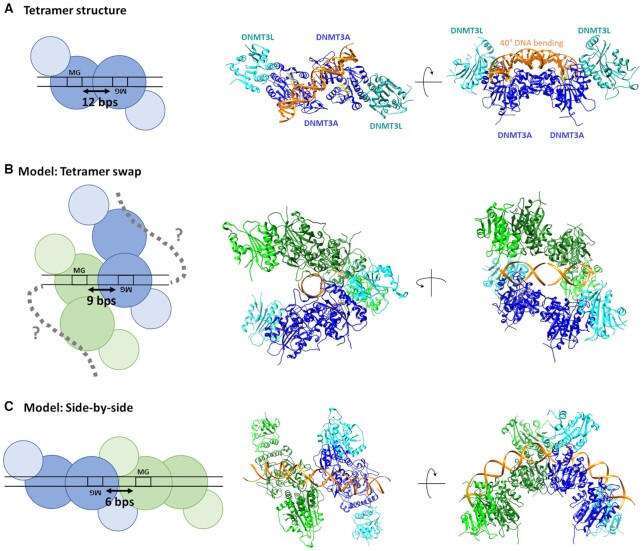
Figure 8.
Schematic pictures, structural snapshots and models explaining MW and WM co-methylation at difference distances. (A) MW co-methylation at 12 bp distance is in agreement with the crystal structure of DNMT3A/3L (6BRR) (22). The two DNMT3A subunits in the heterotetramer are colored blue, DNMT3L is colored cyan, DNA orange and AdoHcy is shown in yellow. (B) MW co-methylation at shorter distances can be explained, if two adjacent tetramers interact with the DNA with only one DNMT3A subunit leading to a tetramer swap of the DNA. The model was generated by using two structures of DNMT3A/3L complexes with two short DNA substrates each bound to one DNMT3A subunit (6F57) (22). The DNA molecules of both tetramers were fused leaving a distance of 9 bp between the CpG sites. (C) WM co-methylation can be explained by two adjacent DNMT3A tetramers interacting with the DNA side-by-side. The model was generated by using two DNMT3A/3L structures with long DNA substrates bound to both DNMT3A subunits (6BRR (22)). The DNA molecules were aligned leaving a distance of 7 bp between the CpG sites.