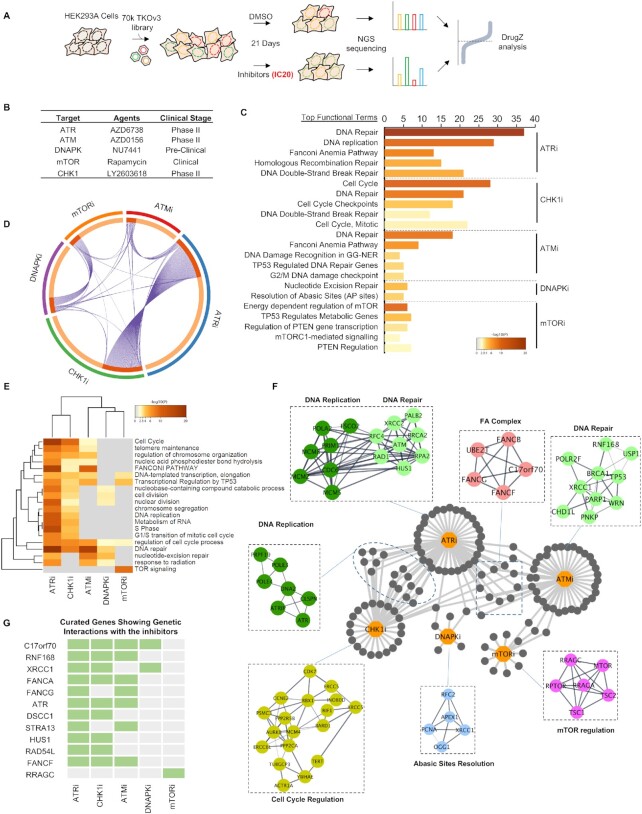
Figure 1.
Genome-Wide CRISPR Screens with DDR Inhibitors in HEK293A Cells. (A) Schematic representation of the workflow for CRISPR screens performed in HEK293A cells. (B) Inhibitors used in the screens and their status in clinical trials. (C) Functional term analysis of the synthetic lethality genes identified in these CRISPR screens. The top five functional terms (REACTOME) enriched for each inhibitor are listed here. The numbers represent the number of genes enriched for each functional term. Different colors represent P values (Benjamini-Hochberg false discovery rate corrections). The functional term analyses were conducted with Metascape (https://metascape.org). (D) Circos graph of the synthetic lethal/sick genes for each inhibitor. The genes shared by different inhibitors are marked with lines. (E) Heatmap representation of the five CRISPR screens with different inhibitors. The top functional terms for each group were listed and the distances between the different groups were calculated by Metascape (http://metascape.org). Gray coloring means that the functional term was not enriched in the indicated group. Other colors represent P values (Benjamini-Hochberg false discovery rate corrections). (F) Network of identified genes in the top enriched functional terms in each group. Nodes were marked out and labeled according to their functions in different biological processes. (G) Fingerprint plot of highlighted genes across the DDR-inhibitor screens. The genes shared by different groups are marked.