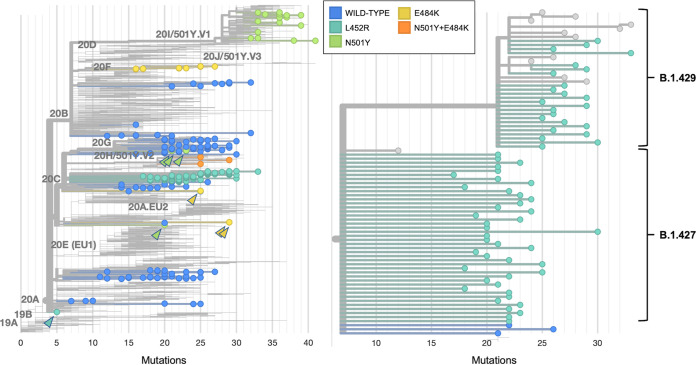
FIG 3.
Nextclade phylogenetic tree of 2,130 SARS-CoV-2 genomes, including 225 of the 229 sequenced genomes from this study and 1,905 genomes from the Nextstrain global reference tree (left). There was one genome excluded because the quality was insufficient to assign a clade. The 225 included genomes are colored by RT-qPCR genotyping result, with each circle representing a sequenced genome. Branch length corresponds to nucleotide divergence. Arrowheads point to mutations identified either sporadically or in rare lineages. Isolates with N501Y mutations belong predominantly to clade 20I/501Y.V1 and lineage B.1.1.7, though sporadic mutations are noted in clade 20G lineage B.1.2 (top three green arrowheads) and clade 20A lineage B.1.480 (bottom green arrowhead). In addition to identifying the clade 20B lineage P.2 variant of concern, E484K RT-qPCR also identified a clade 20C lineage B.1.526 variant of interest (top two yellow arrowheads), as well as a clade 20A proposed lineage B.1 (bottom yellow arrowhead). Isolates with the L452R mutation (turquoise) belong predominantly to clade 20C and lineages B.1.429 and B.1.427 (right). A sporadic L452R mutation was noted within one sample belonging to clade 20A lineage B.1 (turquoise arrowhead). PANGO lineage names and Nextstrain clade names were assigned as of 5 April 2021.