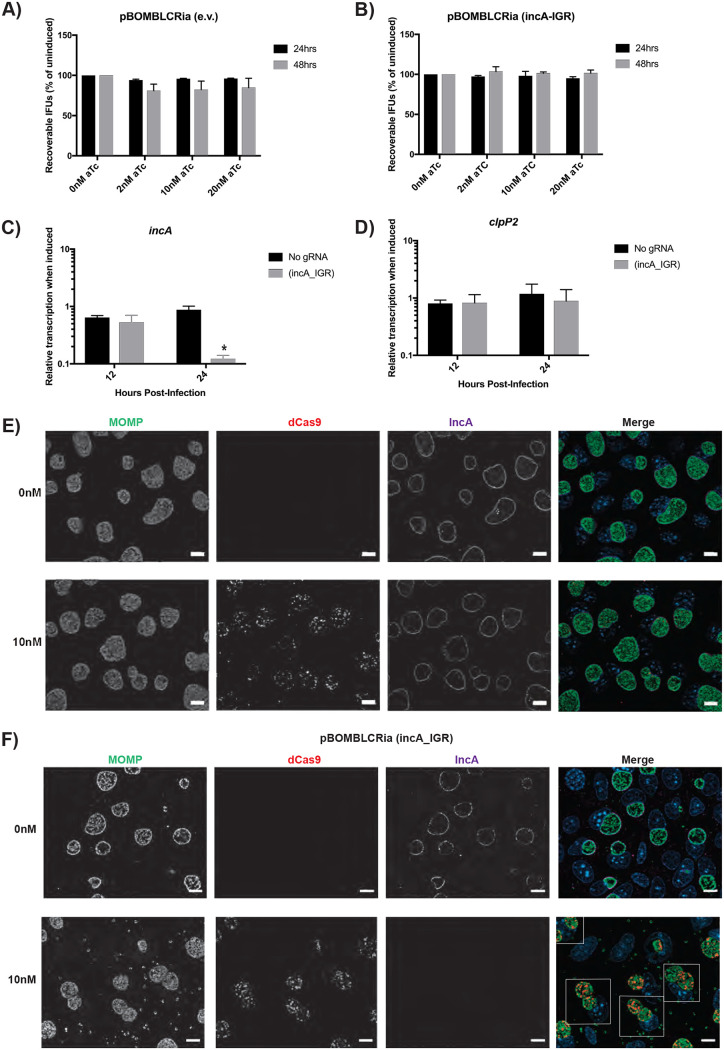
FIG 2.
dCas9 induction effectively represses IncA expression without off-target growth defects. (A and B) McCoy cells were infected with dCas9 transformants with or without an incA-targeting guide RNA (gRNA). Transformants were induced at 4 hours postinfection (hpi) with the various concentrations of aTc and allowed to proceed for 24 hpi, at which time the cells were harvested. The elementary bodies (EBs) were infected onto a fresh monolayer of cells, and this secondary infection was allowed to proceed for 24 h, at which time the inclusion-forming units (IFUs) were counted. No changes between the induced and uninduced control were significant. (C and D) Transformants were induced at 8 hpi using 10 nM aTc. RNA and DNA were isolated at 12 and 24 hpi for quantitative PCR (qPCR). Quantified cDNA was normalized to gDNA and is expressed as induced relative to uninduced samples. Student’s t test was used to compare induced to uninduced samples following log10 transformation, *, P < 0.05. No differences in the clpP2 transcripts were significant. (E and F) Transformants were induced at 4 hpi with 10 nM aTc and allowed to proceed until 24 hpi. Cells were then fixed and stained using primary antibodies to major outer membrane protein (MOMP), ddCpf1 (dCas12), and IncA. Boxes outline cells containing multiple inclusions. All images were acquired on an Axio Imager.Z2 microscope with ApoTome.2 at ×100 magnification. Bars, 10 μm.