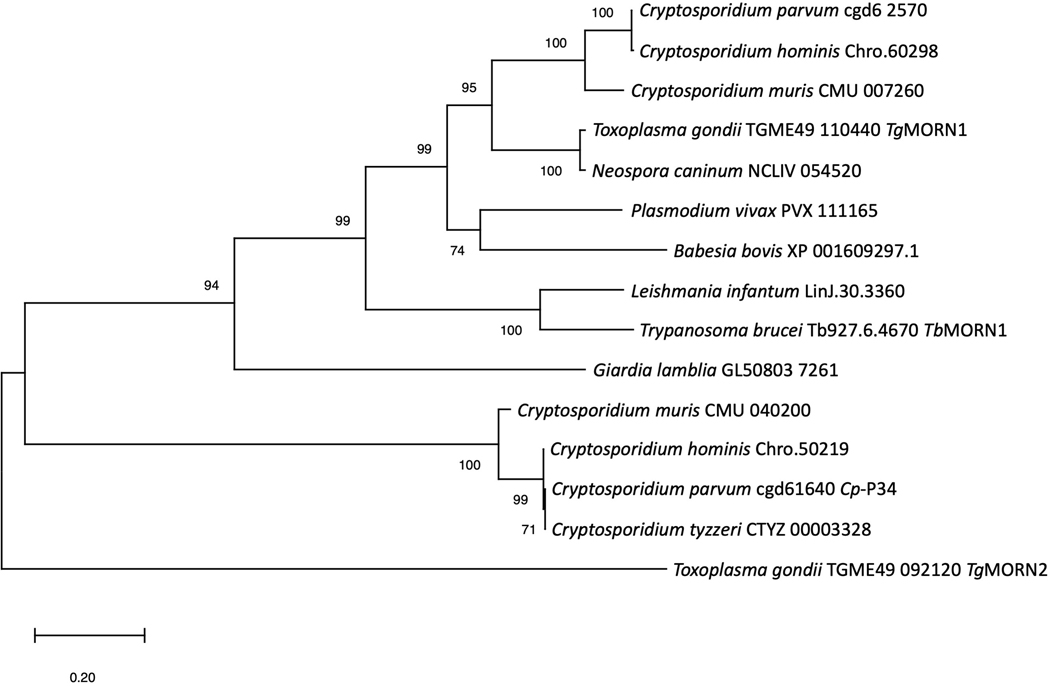
Fig. 6.
Maximum likelihood phylogenetic tree of selected orthologs of P34 of Cryptosporidium parvum (Cp-P34) orthologs. Cp-P34 coded by the cgd5_1640 gene and its close orthologs are unique and cluster away from the group of orthologs of TgMORN1 of Toxoplasma gondii and TbMORN1 of Trypanosoma brucei, including those of Cryptosporidium spp. The evolutionary history was inferred by using the Maximum Likelihood method with 500 bootstrap replications and JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (−6163.19) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 15 amino acid sequences. There was a total of 383 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018; Stecher et al., 2020).