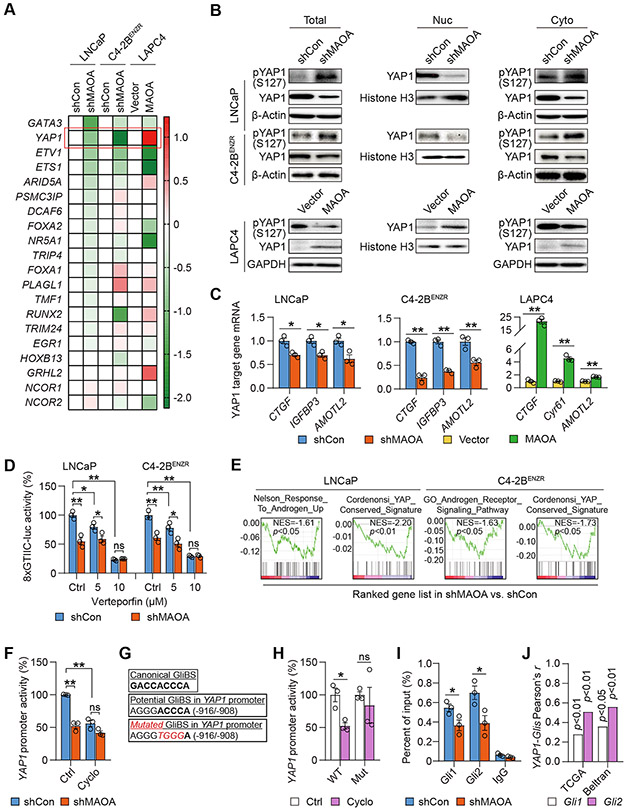
Figure 3. MAOA activates AR-interacting YAP1 in a Gli1/2-dependent manner.
(A) qPCR-based heatmap depicting differential expressions of AR-interacting transcription factors and cofactors in the indicated cell pairs. Log2 scale was used to indicate relative gene expression from an average of 3 replicates, with gene expression in controls set as 1. (B) Western blot of YAP1 and phospho-YAP1 (S127) in total cell lysates along with nuclear and cytoplasmic fractions of the indicated cells. (C) qPCR of YAP1 target genes in the indicated control and MAOA-KD cells (n=3). (D) Determination of YAP1-responsive 8xGTIIC-luc activity by verteporfin (24 hrs) in the indicated control and MAOA-KD cells (n=3). (E) GSEA of the indicated gene sets for the comparisons of MAOA-KD versus control LNCaP and C4-2BENZR cells. (F) Determination of YAP1 promoter activity by cyclopamine (20 μM, 48 hrs) in control and MAOA-KD LNCaP cells (n=3). (G) Sequences of the canonical GliBS (top), a putative GliBS in YAP1 promoter (middle), and introduced point mutations (bottom, italic and red) to inactivate the GliBS, with the YAP1 TSS set as +1. (H) Determination of WT and mutant (Mut) YAP1 promoter activity by cyclopamine (20 μM, 48 hrs) in 293T cells (n=3). (I) ChIP-qPCR of Gli1 and Gli2 occupancy at the GliBS-centric YAP1 promoter in control and MAOA-KD LNCaP cells (n=3). (J) Pearson correlation analysis of YAP1-Gli1/Gli2 in the TCGA (n=498) and Beltran (n=49) datasets. Data represent the mean ± SEM. *p<0.05, **p<0.01; ns, not significant.