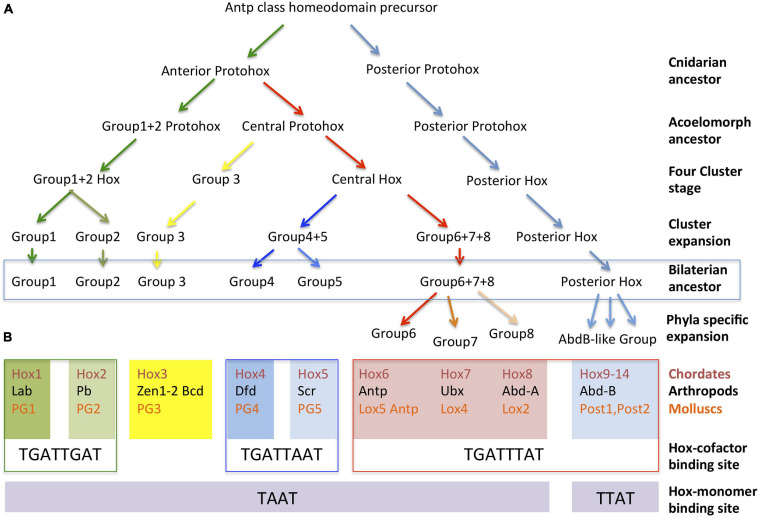
FIGURE 2.
Evolution of Hox protein clusters and correlation with their DNA binding preferences. (A) Inferred cluster evolution from sequential tandem duplications of an ancient Antp class homeobox gene. Three tandem duplications gave rise to a four-gene ProtoHox cluster. Duplication of the full ProtoHox cluster gave rise to the ParaHox cluster (not shown) and the Hox cluster proper. The duplication of specific genes in the four-gene Hox cluster resulted in its expansion in the common ancestor that gave rise to all bilateral animals. Phyla specific duplications resulted in the final expansion that resulted in the 10–15 Hox clusters found in modern species. (B) Classification of the Chordate, Arthropod, and Mollusc Hox proteins into paralogy groups according to their evolutionary relationships as inferred from their sequence similarities. Each of the seven groups is aligned over their preferred DNA binding sites either when in complex with TALE cofactors or as monomers. For simplicity, the Hox6-Hox8 genes have been organized in columns aligned with particular arthropod and mollusc equivalents, even though direct homology for genes in the column has not been demonstrated, see text.