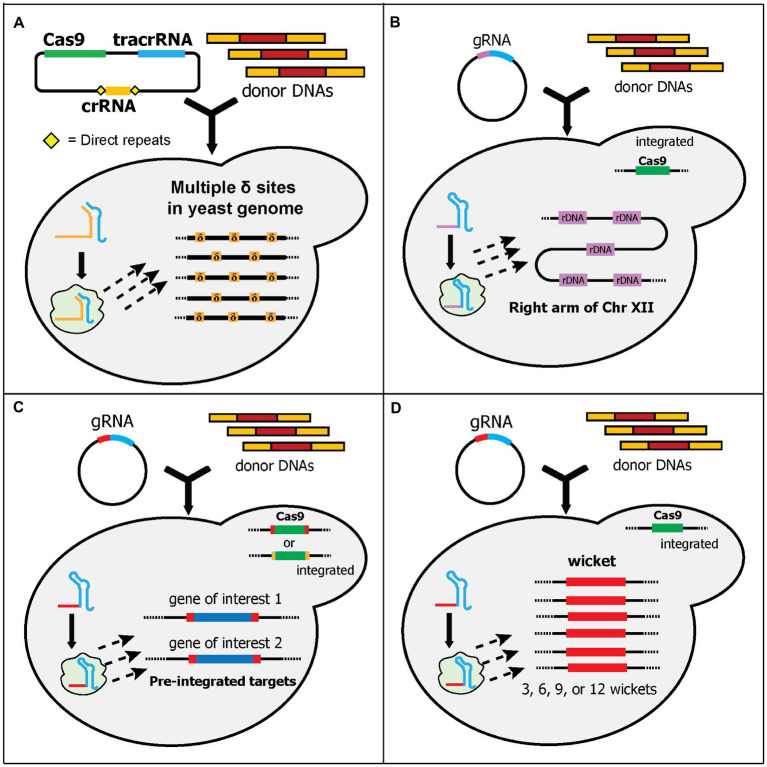
Figure 4.
Schematic diagram of multiplex gene-editing pre-defined sequences in the genome using a single gRNA (part 1). (A) Shi et al. used δ sites of Ty elements in the yeast genome as the target. More than 30 δ sites (δ, orange boxes) are scattered across all chromosomes in the yeast genome. Therefore, multiple copies of donor DNA can be integrated. Note that they used a separate crRNA and tracrRNA plasmid. (B) Wang et al. targeted ribosomal DNA (rDNA, purple boxes) for integration sites. rDNA is located at the right arm of chromosome XII. Around 100–200 copies of rDNA can be found in the yeast genome. Cas9 was integrated into the genome. (C) Finnigan and Thorner generated synthetic target sequences (red or orange boxes) that flanked the genes-of-interest (GOIs) and Cas9, which was integrated into the yeast genome. The gRNA cleaves the sequences and causes DSBs at the 5' and 3' ends of GOIs and/or Cas9, removing them from the genome. Concurrently, the donor DNAs can be integrated at those sites. Note that although different gRNA sequences can be used, only one gRNA sequence (red box) is shown. (D) Hou et al. generated “wicket” sequences (red boxes) which contains a 23-bp synthetic target flanked by 5' and 3' 50-bp synthetic homology arms. Wickets and Cas9 were integrated into the yeast genome. Yeast strains with 3, 6, 9, or 12 wickets were created. Therefore, multi-copies of donor DNA can be integrated in one transformation.