Figure 5.
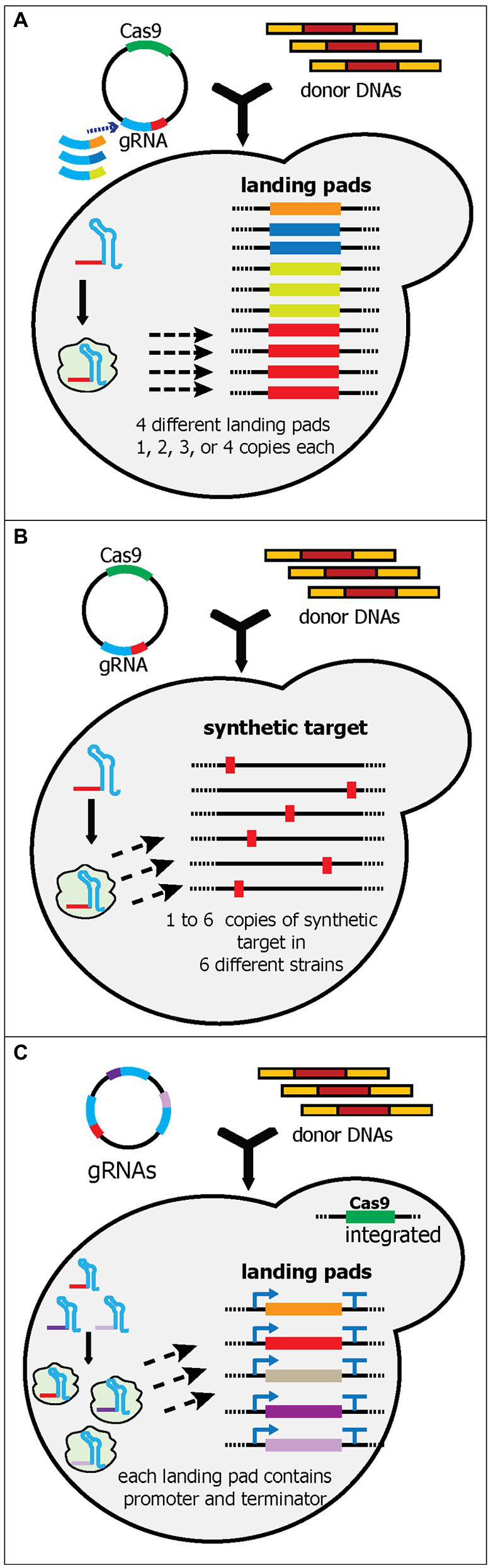
Schematic diagram of multiplex gene-editing pre-defined sequences in the genome using a single gRNA (part 2). (A) Bourgeois et al. tested and selected various landing pads sequences. Like wickets, the landing pads contain a 23-bp synthetic target flanked by 280-bp 5' and 3' synthetic homology arms. Four unique landing pads (orange, dark blue, light green, and red boxes) were integrated into 10 characterized loci in the yeast genome. Each landing pad had a different copy number in the genome; i.e., landing pad 1 has one, and landing pad 2 has two. Note that four different plasmids were created, each of which targeted a different landing pad. Only one landing pad gRNA (red) was shown in the yeast cell. (B) Baek et al. generated and integrated a 23-bp synthetic sequence (red boxes) into one to six characterized loci in the yeast genome. Six yeast strains were constructed, each of which had from one to six synthetic sequence(s). Therefore, up to six genes could be integrated simultaneously. (C) Qi et al. created a system called PCR & Go. Up to eight 23-bp synthetic sequences flanked with unique sets of promoters and terminators were integrated into the yeast genome. The gRNA plasmid contains multiple gRNA cassettes instead of a single gRNA. However, donor DNA preparation can be simplified as unique sets of promoters and terminators had already been integrated into the genome.
