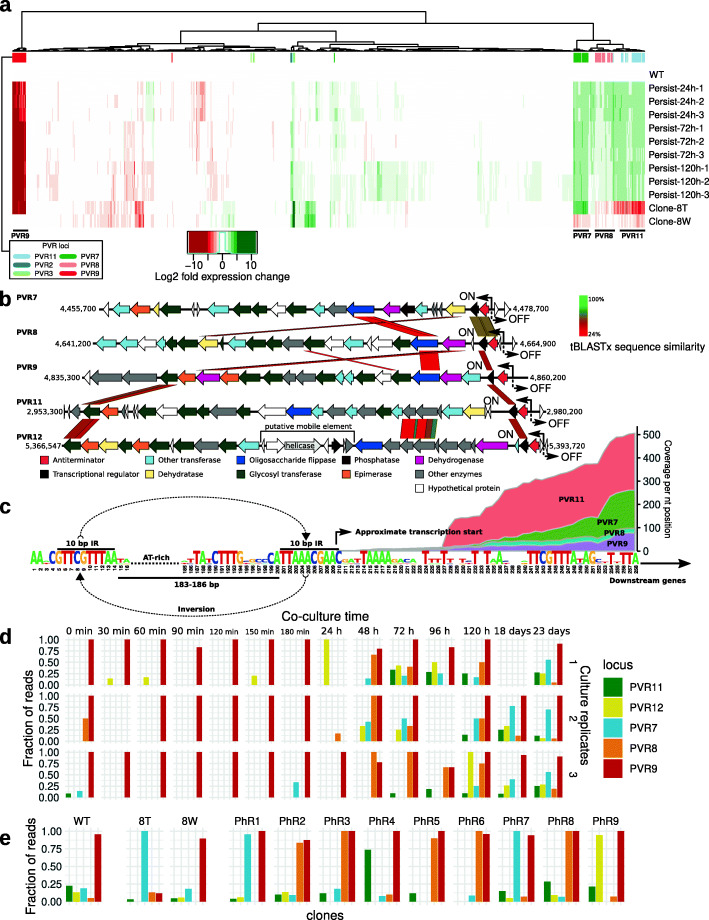
Fig. 3.
Structure and transcriptional control of five phase-variable capsular polysaccharide (CPS) operons (PVR7, PVR8, PVR9, PVR11, PVR12) in B. intestinalis APC919/174. a Host transcriptional response in a B. intestinalis APC919/174-ΦcrAss001 serial broth co-culture (24-120h in vitro persistence) experiment, analyzed using RNAseq (values are log2 fold gene transcript expression change relative to the un-infected control from two independent experimental runs); clone-8T, phage-resistant derivative of the parental strain; clone-8W, spontaneous phage-sensitive revertant clone; only genes with significantly changed expression (p < 0.01 in DESeq2, n = 826) are shown; genes, associated with phase-variable genomic regions (PVR) are marked with color bars on top; b structure of phase variable operons, encoding five different CPS; protein sequence homologies (tBLASTx) between gene products are shown as colored parallelograms; two-sided arrows and ON/OFF labels mark positions of invertible promoters; c consensus sequence of the regions flanking invertible promoters (ON orientation) in CPS biosynthesis loci; approximate transcription start was identified though analysis of RNAseq data; stacked area charts show RNAseq read coverage of regions adjacent to the promoter in four CPS loci; d, e fraction of concordantly-aligned Illumina read pairs supporting orientation of invertible promoters in ON direction; WT, wild type strain; clones 8T, PhR1-9, phage-resistant derivatives; 8W, phage-sensitive revertant; P18d/P23d, samples in e were taken from the long-term in vitro persistence experiment (also see Additional file 1: Fig. S3 for similar data on the long term in vivo persistence)