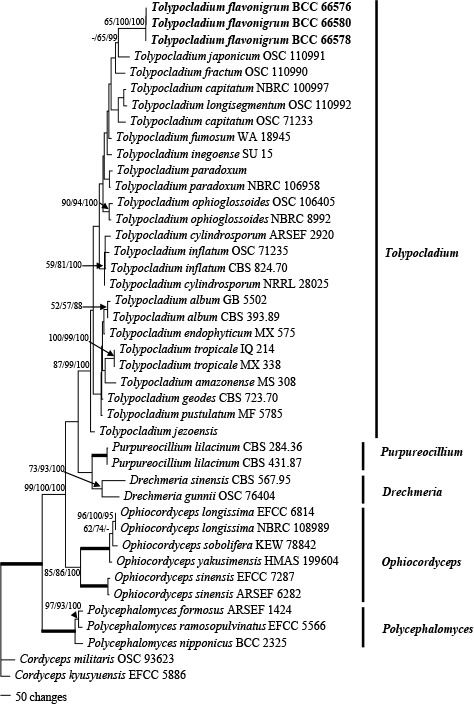
Phylogenetic tree with T. flavonigrum constructed from a combined dataset comprising LSU, tef1 and rpb1. The phylogenetic tree was analysed using Maximum parsimony (MP), Maximum likelihood (ML) and Bayesian inference. The MP analysis was conducted on the combined data set using PAUP v. 4.0b10 (Swofford 2003), adopting random addition sequences (100 replications), with gaps being treated as missing data. A bootstrap (BP) analysis was performed using the maximum parsimony criterion in 1 000 replications. The ML analysis was run with RAxML-VI-HPC2 v. 8.2.12 (Stamatakis 2014) under a GTR model, with 1 000 bootstrap replicates. Bayesian phylogenetic inference was calculated with MrBayes v. 3.2.6 (Ronquist & Huelsenbeck 2003), with 5 M generations and under the same model. Numbers at the significant nodes represent MP bootstrap support values/RAxML bootstrap support values/Bayesian posterior probabilities (BPP) times 100. Thickened lines in the tree represent 99–100 % bootstrap support values and 99–100 BPP.
