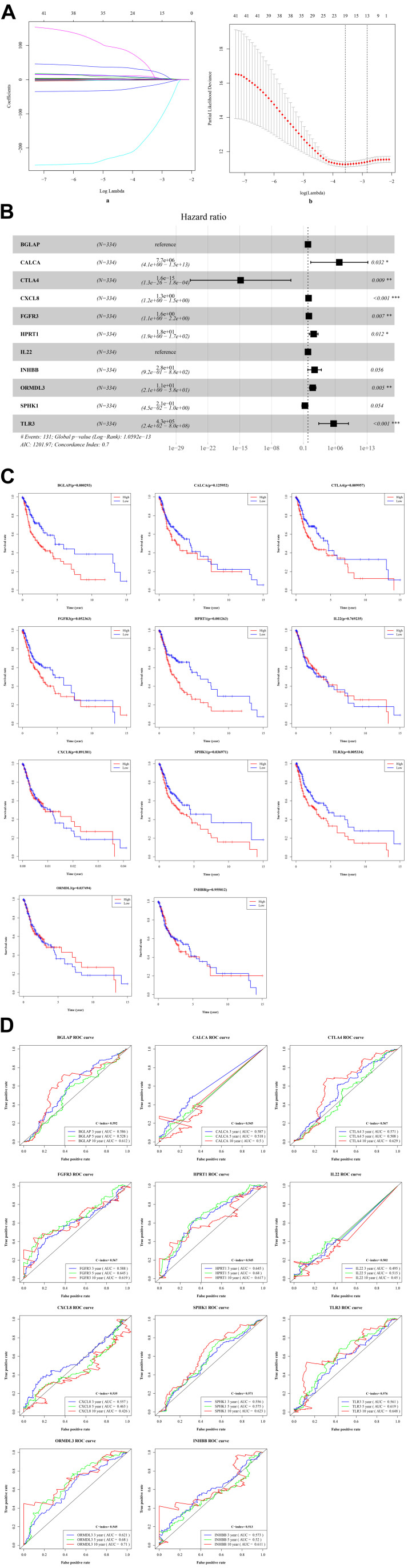
FIGURE 3.
The identification of ISGs with significant prognostic values in OSCC. (A) Demographic and clinical feature selection using the LASSO binary logistic regression model. (a) LASSO coefficient profiles of the 42 gene features. A coefficient profile plot was produced against the log(lambda) sequence. All weights converge toward zero as the penalty parameter increases. Vertical line was drawn at the value selected using fivefold cross-validation, where optimal lambda resulted in five features with non-zero coefficients. (b) Optimal parameter (lambda) selection in the LASSO model used fivefold cross-validation via minimum criteria. The partial likelihood deviance (binomial deviance) curve was plotted versus log(lambda). Dotted vertical lines were drawn at the optimal values by using the minimum criteria and the 1 standard error (SE) of the minimum criteria (the 1-SE criteria). (B) The forest plot with hazard ratio (HR) for the 11 risk ISGs. HRs above one indicates that a gene is positively associated with the event probability and thus negatively with survival time. The box size is based on precision, and the x-axis has a logarithmic scale. A bigger box size represents a more precise confidence interval (95% CI). (C) The Kaplan–Meier curves of the 11 risk ISGs. Red lines represent high expression group, while blue lines represent low expression group. The upper line indicates the higher survival rate, while the lower line indicates the lower survival rate. (D) The time-dependent receiver operating curve (ROC) is generated for the survival prediction of 11 risk ISGs. The time-dependent (3-, 5-, and 10-year) area under curve (AUC) and C-index for each risk ISG are respectively labeled in the lower right corner of each gene’s ROC curve.