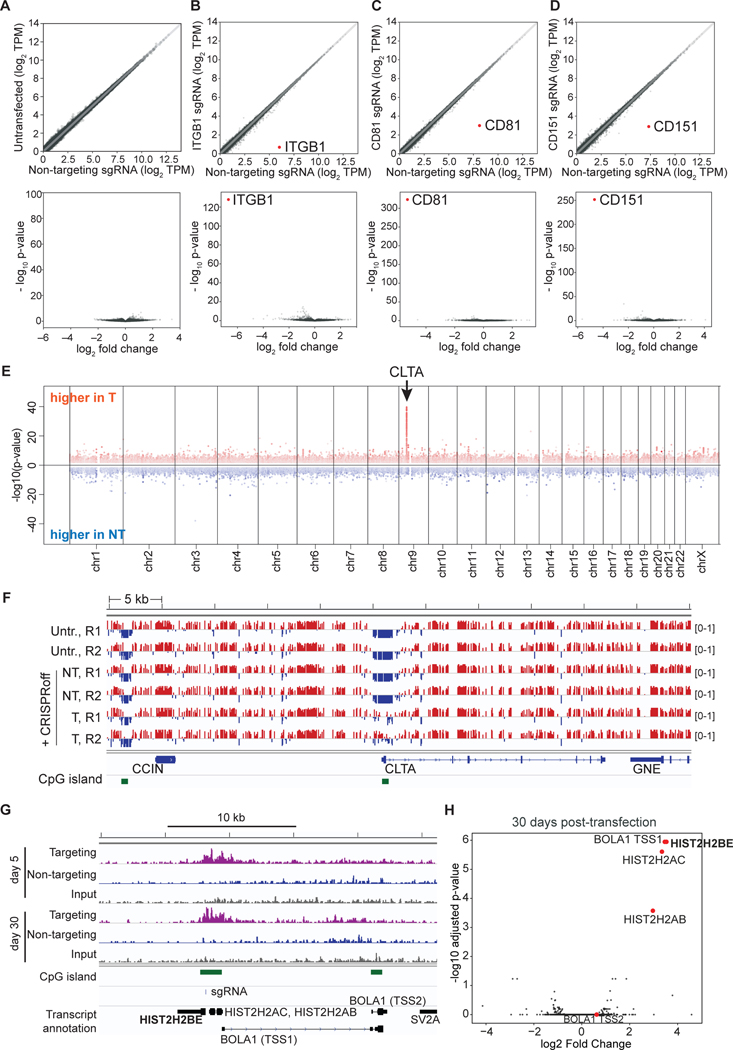
Figure 2. Highly specific and robust transcriptional silencing by CRISPRoff.
(A–D) RNA-seq plots of HEK293T cells transfected with CRISPRoff and non-targeting (NT) sgRNAs compared to sgRNAs targeting (B) ITGB1, (C) CD81, or (D) CD151. A comparison of untransfected cells and CRISPRoff with NT sgRNA is shown in (A. The volcano plots (bottom) display the targeted genes as the most significantly repressed transcripts globally. The data are representative of the average of two independent replicates.
(E) A Manhattan plot displaying differentially methylated CpGs between cells treated with CRISPRoff and CLTA-targeting or NT sgRNAs (30 days post-transfection) analyzed by WGBS. Red dots represent CpGs that gained DNA methylation in targeting sgRNA cells and blue dots represent CpGs that gained DNA methylation in NT sgRNA cells. The arrow denotes the genomic position of CLTA.
(F) A comparison of CpG methylation along a 55 kb window that includes the CLTA locus. Tracks labelled ‘Untr.’ represent untransfected cells; the ‘NT’ tracks represent cells transfected with CRISPRoff and non-targeting sgRNA; the ‘T’ tracks represent cells transfected with CRISPRoff and targeting sgRNA. R1 and R2 represent two technical replicates. Red marks represent methylated (beta-value >0.5) and the blue marks represent unmethylated (<0.5) CpG dinucleotides. CpG islands are shown in green.
(G) A comparison of H3K9me3 ChIP-seq signal across the H2B gene in cells transfected with CRISPRoff and H2B-targeting (purple) or NT sgRNAs (blue) taken at 5 days and 30 days p.t. The sgRNA binding site is denoted along with the CpG islands and neighboring genes. The BOLA1 gene contains two annotated transcriptional start sites, labeled TSS1 and TSS2.
(H) Volcano plot comparing H3K9me3 ChIP-seq data between CRISPRoff transfected with either H2B-targeting or NT sgRNAs. Red dots highlight the genes proximal to the H2B target.