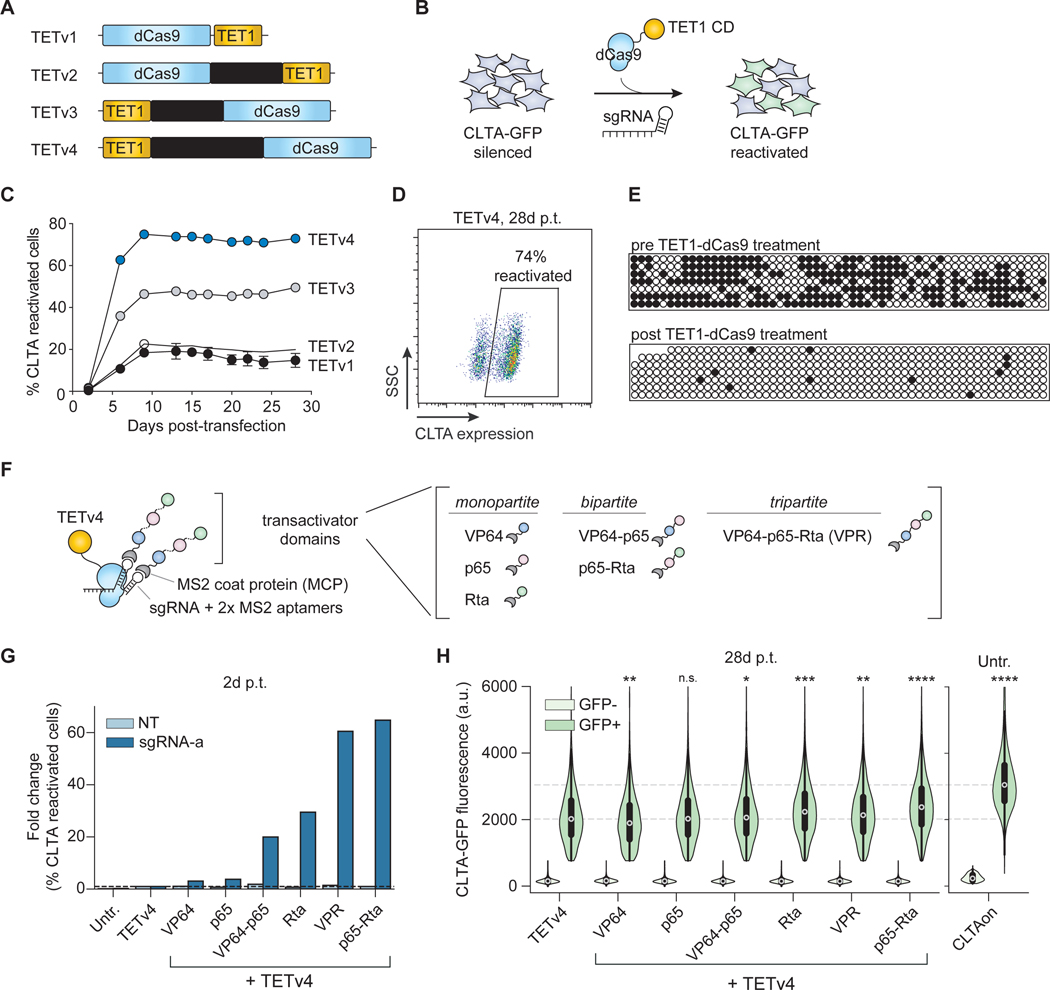
Figure 3. CRISPRon reverses silenced genes by combining DNA demethylation and transcriptional activators.
(A) A schematic of the four TET1 catalytic domain fusions to dCas9 (TETv1-v4) that were tested for reactivation of CRISPRoff-silenced genes.
(B) CRISPRoff-silenced CLTA-GFP cells were transfected with plasmids encoding TETv1–4 and targeting or NT sgRNAs to assess reactivation.
(C) A time course of CLTA reactivation after transfection of each of the four TET fusions in (A). The mean values were measured from three independent experiments. Error bars represent SD.
(D) A representative flow cytometry plot of CLTA reactivation measured at 28 days p.t. of TETv4 and targeting sgRNAs.
(E) Bisulfite-PCR analysis of the CLTA CGI after reactivation by TET1v1 shows high levels of demethylated CpG cytosines (white circles) compared to CRISPRoff-silenced cells.
(F) A schematic of the TETv4 and transactivator ribonucleoprotein complex mediated by a sgRNA encoding two MS2 RNA aptamers. Transactivator domains include monopartite, bipartite, and tripartite architectures of VP64, p65, and Rta.
(G) Fold change in the fraction of CLTA-GFP reactivated cells compared to TETv4 alone, measured two days p.t. The data are calculated from the mean of eight technical replicates from three independent experiments.
(H) Comparison of the expression of CLTA-GFP in single cells, measured by cytometry 28 days p.t. with CRISPRon. The data are aggregated from three technical replicates. * p value < 1e-4, ** p value < 1e-20, *** p value < 1e-100, **** p value = 0, relative to the GFP positive population in the TETv4 condition by the Wilcoxon rank-sum test. Unsilenced CLTA-GFP cells are provided as a benchmark for wild type expression levels.