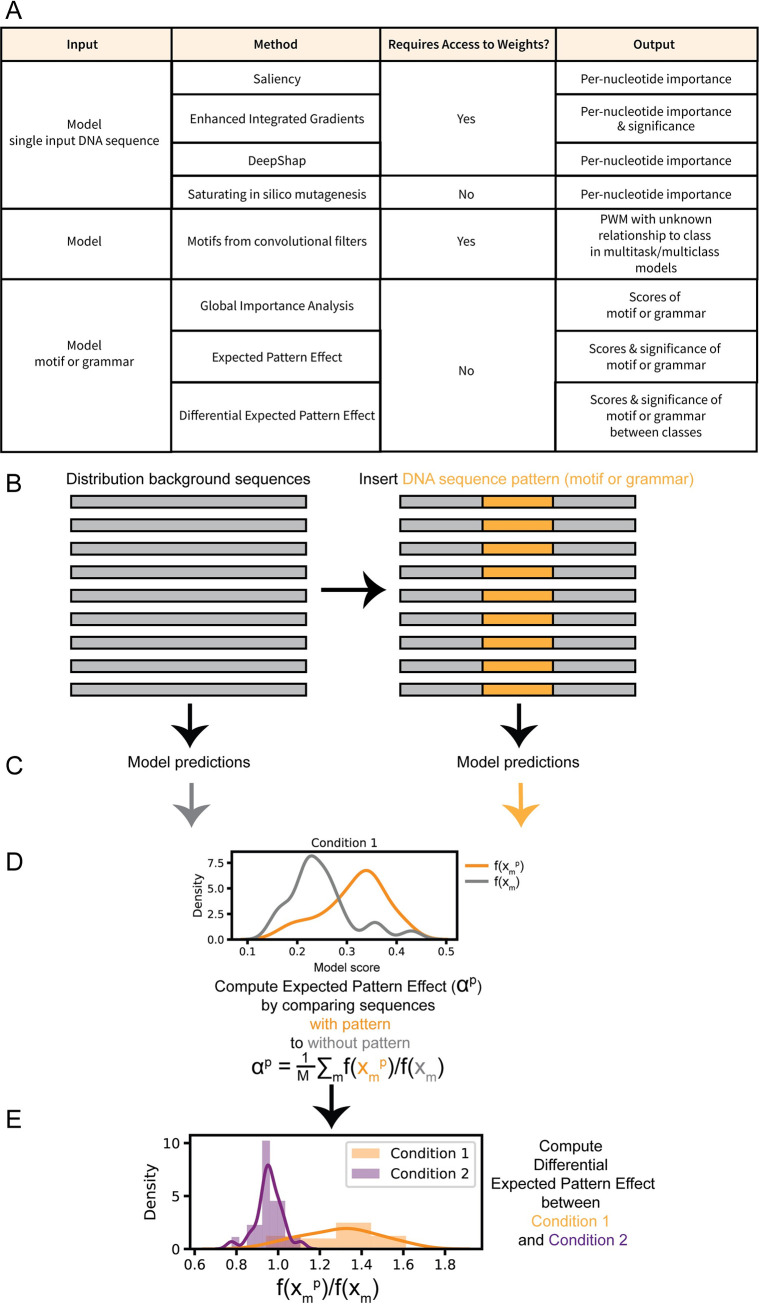
Fig 1. Expected Pattern Effect and Differential Expected Pattern Effect determine the significance of feature patterns such as transcription factor motifs or transcription factor grammar for multitask models.
(A) Comparison of interpretation methods shows 3 categories of interpretation methods for deep learning models based on input. Some methods operate on an individual sequence input, model pair, others turn learned convolutional filters into DNA sequence motif position weight matrices and then must assign class relevance post-hoc, or methods Global Importance Analysis and Expected Pattern Effect score the overall class importance of DNA sequence patterns such as transcription factor motifs or grammar such as combinations or spacing of transcription factor motifs. (B) Computation of Expected Pattern Effect and Differential Expected Pattern Effect starts by selection of a sequences that represent a “background” from the natural distribution of sequences. A pattern of interest either a transcription factor or combination or spacing of transcription factors is artificially inserted into each background sequence. (C) Model predicts functional signal for “background” sequences and sequences with inserted transcription factor motif. In our case we use a DeepAccess model, but principles for computing EPE and DEPE can be applied to any multi-task classification model. (D) Average ratio between scores of background sequences with pattern of interest (in orange) and without pattern of interest–i.e. background sequences (in grey) determines the Expected Pattern Effect. (E) Differential Expected Pattern Effect can be computed by comparing Expected Pattern Effect between different classes–i.e. conditions or cell types.