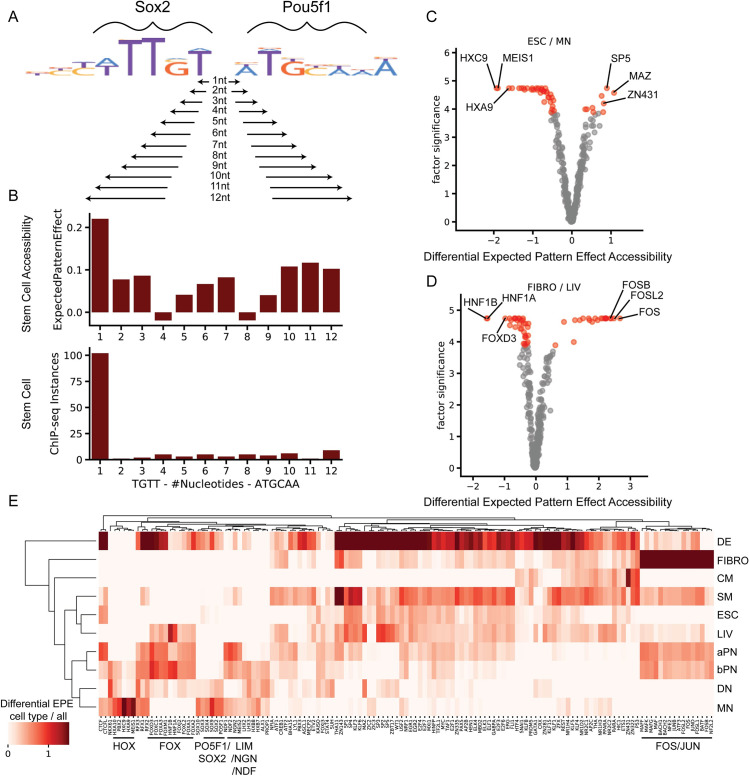
Fig 3. Expected Pattern Effect and Differential Expected Pattern Effect identify preferential spacing and cell type-specific transcription factor activity from DeepAccess models trained on chromatin accessibility data from ten cell types.
(A) Testing all possible spacing between Sox2 transcription factor and Pou5f1 (Oct4) transcription factor motifs input as patterns to compute Expected Pattern Effects from a DeepAccess model. (B) DeepAccess Expected Pattern Effect of Sox2 and Pou5f1spacing on predicted stem cell accessibility (top) is consistent with highest score for preferred spacing of Sox2 and Pou5f1 motifs from ChIP-seq binding data of Sox2 and Pou5f1 co-binding events. (C) Differential EPE of transcription factor motifs between stem cells (ESC) and motor neurons (MN) identifies known MN-specific transcription factors such as Hox TFs and Meis. Scatter plot y-axis is unadjusted rank ratio statistic, x-axis is the Differential Expected Pattern Effect (log2 fold change in Expected Pattern Effect between stem cells and motor neurons), with each point representing one of 356 HOCOMOCOv11 transcription factor motifs. Red points are significant (p < 0.05) transcription factor motifs under Bonferroni multiple hypothesis correction. (D) Differential EPE of transcription factor motifs between fibroblasts (FIBRO) and liver cells (LIV) identifies liver-determining factors such as Hnf1 and Fox transcription factors. (E) Transcription factors showing significant cell type-specific EPEs for each of the ten cell types relative to all other cell types. Heatmap intensity indicates the log2 fold change in Expected Pattern Effect between each cell type and the average of all other cell types.