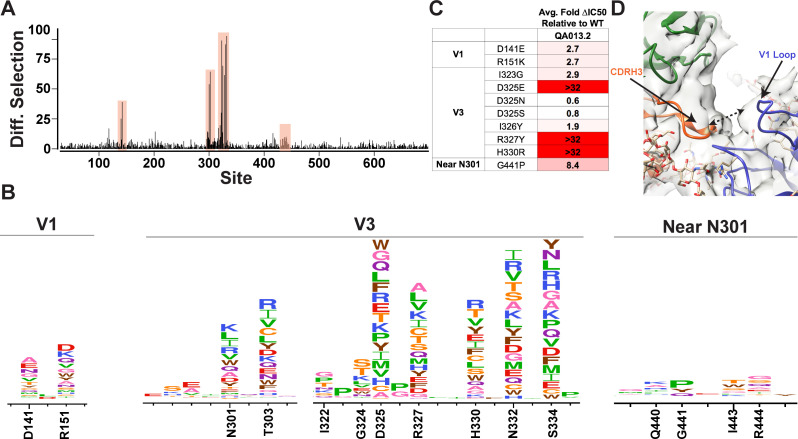
Figure 7. Mutational antigenic profiling of QA013.2 identifies unique viral escape mutations.
(A) Line plot showing the median positive differential selection across the mutagenized portion of BG505.W6.C2 T332N Env. (B) Logoplots showing the mutation-level enrichment for areas of escape in A. (C) Table of average fold change in IC50 for each BG505.W6.C2 T332N viral escape mutant relative to the wildtype BG505.W6.C2 T332N pseudovirus as tested in the TZM-bl neutralization assay. Darker red colors indicate a larger fold change in average IC50. In vitro neutralization data are representative of three independent replicates. (D) Heavy chain CDR3 loop is within 8 Å distance to the V1 loop when measured between the Cα chains. See also Figure 7—figure supplements 1 and 2 and Figure 7—source data 1.