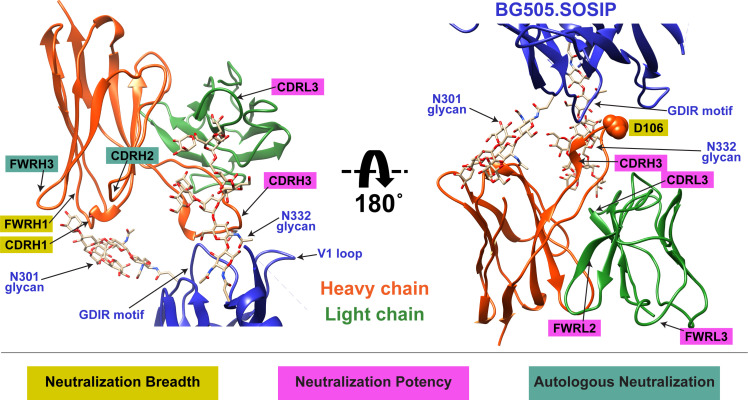
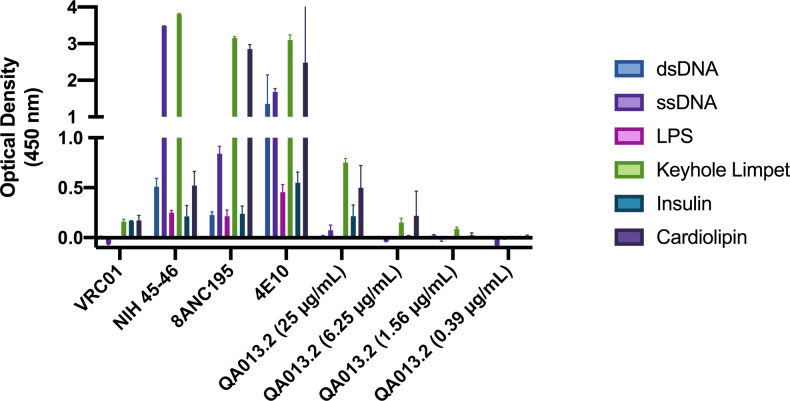
Figure 9. Summary of QA013.2 determinants of HIV neutralization breadth, potency, and autologous neutralization.
Ribbon structure of QA013.2 Fab variable domain complexed to BG505.SOSIP.664 trimer in two different orientations are shown. Env structure is colored in blue with protruding glycans at sites N301 and N332 shown as stick models, Fab heavy chain in orange, and Fab light chain in green. Key regions of the Fab structure that confer breadth (yellow) or potency (pink) are labeled and colored categorically. Teal colored regions (CDRH2 and FWRH3) confer autologous neutralization. Important Env epitopes of interaction such as the linear GDIR motif and V1 loop are also labeled. See also Figure 9—figure supplement 1.