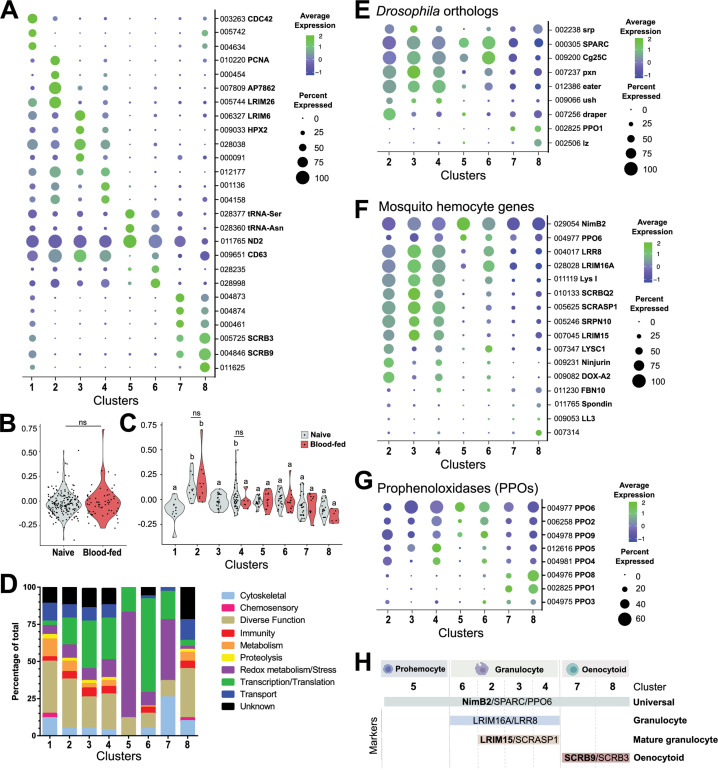
Figure 2. Comparative analysis of mosquito immune cells.
(A) Marker gene expression displayed by dot plot across cell clusters. Dot color shows levels of average expression, while dot size represents the percentage of cells expressing the corresponding genes in each cell cluster. (B) Violin plot of cell cycle genes (GO::0007049) displayed as the difference in average gene expression levels between the cell cycle gene set in cells under naïve and blood-fed conditions. (C) Similar comparisons of cell cycle genes were examined in individual cell clusters and under naïve and blood-fed conditions where possible. For B and C, positive numbers indicate higher levels of cell cycle gene expression compared to the random set for that physiological condition or cell cluster. (D) Gene ontology (GO) analysis of genes expressed in >80% cells within each respective cluster. Heat maps of candidate genes to Drosophila hemocyte orthologs (E), described mosquito hemocyte genes (F), or An. gambiae prophenoloxidases (PPOs) (G) to enable the characterization of immune cells form each cell cluster. (H) From these analysis, immune cells cluster were assigned to tentative cell types (prohemocytes, granulocytes, oenocytoids) based on the expression subtype-specific marker expression. Genes in bold are featured prominently in our downstream analysis.
Figure 2—figure supplement 1. Comparisons of immune cell clusters to the An.
Figure 2—figure supplement 2. Comparisons of Cluster 1 to non-hemocyte cell populations.
Figure 2—figure supplement 3. Expression of hemocyte genes across all cell clusters.
Figure 2—figure supplement 4. Expression of mosquito immune genes across cell clusters.

Figure 2—figure supplement 5. Expression of serine protease inhibitors (SRPNs) and CLIP serine proteases across cell clusters.

Figure 2—figure supplement 6. Expression of chemosensory genes across cell clusters.
Figure 2—figure supplement 7. Expression of specific tRNAs across cell clusters.