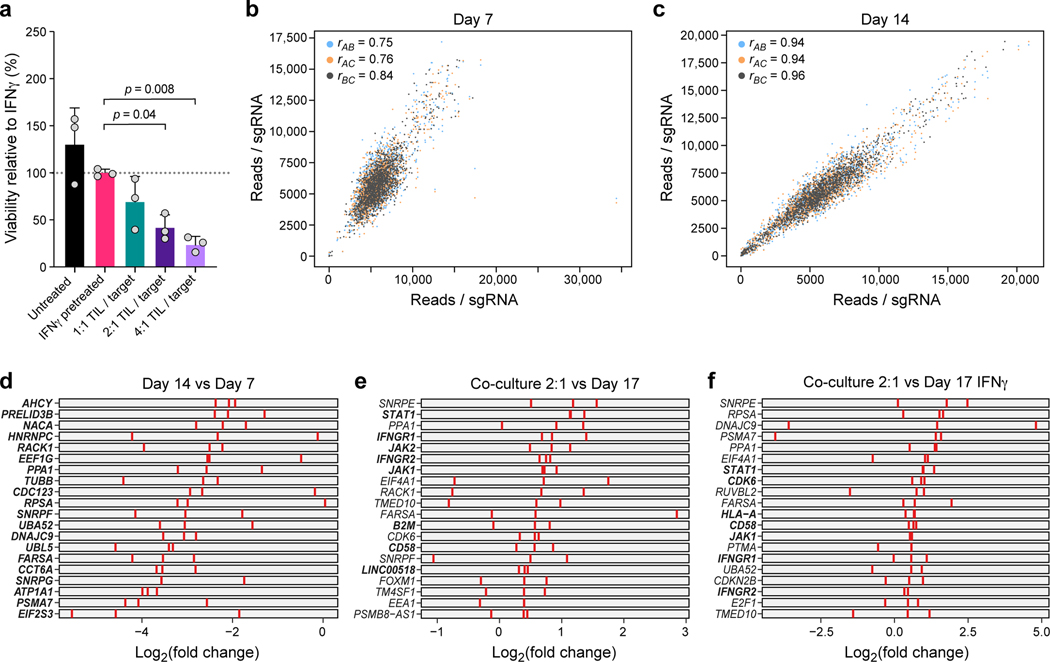
Extended Data Fig. 3. CRISPR/Cas9 viability screen in the co-culture system.
a, Dose-response killing in the co-culture experiment validates target killing range. Percent of surviving cells relative to IFNγ pretreated target cells (y axis) in different co-culture conditions (x axis) from a plate run in parallel to the viability and Perturb-CITE screens, with triplicate wells for each condition. One-way ANOVA with Dunnet post-hoc test. Error bars: Mean±SD. b,c, Screen reproducibility across triplicates. Number of reads detected (x, y axis) for each sgDNA (dots) when comparing each pair within triplicate experiments (color legend) in pre-treated day 7 (b) or day 14 (c). Pearson correlation coefficients are noted in the color legend. d-f, Identification of essential genes and genes affecting resistance to TIL mediated killing. Relative depletion (log2(FC), x axis) for each individual sgDNA (red bar) of the top20 target genes by MAGeCK analysis (rows) (n = 3 sgDNAs/target gene, Methods) on day 7 (without TILs, to recover essential genes, d), day 17 of 2:1 TIL:cancer cells co-cultures, comparing control cells (e), or day 17 of IFNγ treated cells (f, no co-culture). Bold lettering indicates significantly enriched / depleted target.