Abstract
Rod and cone photoreceptors of the vertebrate retina utilize cGMP as the primary intracellular messenger for the visual signaling pathway that converts a light stimulus into an electrical response. cGMP metabolism in the signal-transducing photoreceptor outer segment reflects the balance of cGMP synthesis (catalyzed by guanylyl cyclase) and degradation (catalyzed by the photoreceptor phosphodiesterase, PDE6). Upon light stimulation, rapid activation of PDE6 by the heterotrimeric G-protein (transducin) triggers a dramatic drop in cGMP levels that lead to cell hyperpolarization. Following cessation of the light stimulus, the lifetime of activated PDE6 is also precisely regulated by additional processes. This review summarizes recent advances in the structural characterization of the rod and cone PDE6 catalytic and regulatory subunits in the context of previous biochemical studies of the enzymological properties and allosteric regulation of PDE6. Emphasis is given to recent advances in understanding the structural and conformational changes underlying the mechanism by which the activated transducin α-subunit binds to—and relieves inhibition of—PDE6 catalysis that is controlled by its intrinsically disordered, inhibitory γ-subunit. The role of the regulator of G-protein signaling 9–1 (RGS9–1) in regulating the lifetime of the transducin-PDE6 is also briefly covered. The therapeutic potential of pharmacological compounds acting as inhibitors or activators targeting PDE6 is discussed in the context of inherited retinal diseases resulting from mutations in rod and cone PDE6 genes as well as other inherited defects that arise from excessive cGMP accumulation in retinal photoreceptor cells.
Keywords: Photoreceptor, phosphodiesterase, cyclic GMP, visual transduction, review, retinal disease, retina, rod photoreceptor, cone photoreceptor, visual excitation
Introduction
In the vertebrate retina, the initial events in vision occur in the outer segment organelles of rod and cone photoreceptor cells. It is on the specialized outer segment disk membranes [94] that the proteins responsible for the first steps in the visual signaling pathway reside, including the photoactivatable G-protein coupled receptor (rhodopsin (in rods) or cone opsins) and two peripheral membrane proteins, the heterotrimeric G-protein (transducin) and the photoreceptor cGMP phosphodiesterase (PDE6). Photoisomerization of 11-cis retinal bound to rhodopsin/opsin leads (via activation of transducin) to a rapid activation of PDE6 catalytic activity which reduces the cGMP levels in the outer segment, with the subsequent closure of cyclic nucleotide-gated ion channels in the outer segment plasma membrane and membrane hyperpolarization [112]. PDE6 inactivation is primarily accomplished by the action of several additional membrane-associated proteins, namely Regulator of G-protein Signaling 9–1 (RGS9–1), its obligate partner (G-protein β-subunit Type 5L, Gβ5L), and the integral membrane protein RGS9 Anchoring Protein (R9AP) [7]. The return of PDE6 to its nonactivated state in conjunction with calcium-dependent activation of guanylyl cyclase activity restores cGMP levels to their dark-adapted concentrations [135]. Additional cellular and biochemical mechanisms underlying various adaptational properties of rod and cone photoreceptors (light-dependent protein translocation, post-translational modifications of phototransduction proteins, chromophore regeneration, etc.) will not be covered, except insofar as they directly pertain to the regulation of PDE6.
This review is not intended to be comprehensive, but rather emphasizes recent discoveries on the structure, function, and regulation of this central effector of visual signaling, with applications to potential pharmacological interventions for inherited retinal diseases arising from mutations in PDE6 genes. Because of the challenges in obtaining purified cone photoreceptors for biochemical studies, the better characterized rod phototransduction pathway will be the primary focus of this review.
PDE6 structure
Rod and cone PDE6 constitute one of the eleven families of Class I cyclic nucleotide phosphodiesterases that share a highly conserved C-terminal catalytic domain [31,42]. The PDE6 family is one of five families whose N-terminal regulatory region consists of two tandem GAF domains (so-called due to their occurrence in mammalian cGMP-binding PDEs, Anabaena adenylyl cyclases and E. coli FhlA) that bind cyclic nucleotides and are generally believed to allosterically communicate with the catalytic domain [149,60]. While structurally and pharmacologically most closely related to PDE5 [32], PDE6 has several features that distinguish it from the other PDE families: (1) whereas cone PDE6 catalytic subunits (gene name PDE6C, common name α’-subunit) form a homodimer as is the case for the other ten PDE families, most vertebrates have two homologous rod PDE6 catalytic subunit genes (PDE6A (α-subunit; Pα) and PDE6B (β-subunit; Pβ) that form a catalytic heterodimer (Pαβ); (2) the catalytic activity of rod and cone PDE6 operate at the diffusion-controlled limit for enzyme catalyzed reactions, more than 100-fold greater than the catalytic turnover number for any other PDE family; (3) unlike other PDE families, PDE6 is directly activated upon binding of a G-protein α-subunit; (4) rod and cone PDE6 catalytic activity is controlled by the binding of an inhibitory γ-subunit (Pγ; gene names: PDE6G (rod) and PDE6H (cone; abbreviated as Pγ’)) at the entrance of the enzyme active site; (5) proper protein folding of PDE6 catalytic subunits requires the participation of a photoreceptor-specific chaperone protein, aryl-hydrocarbon receptor interacting protein-like 1 (AIPL1); and (6) PDE6 catalytic subunits are post-translationally modified at their C-termini in a sequential manner by prenylation (farnesyl moiety for Pα, geranylgeranyl group for Pβ and for the cone α’-subunit), proteolysis, and carboxymethylation (previously reviewed in [33,136,54,76,7,139].
Until recently, structural information about the molecular organization of the rod or cone catalytic dimer and association with its two γ-subunits was limited to low resolution structures of rod PDE6 holoenzyme (αβγγ) obtained by negative-stain electron microscopic image analysis [71,70,47], an x-ray structure of the monomeric cone PDE6 GAFa domain (PDBID: 3DBA) [89] and of a chimeric PDE5/PDE6 catalytic domain complexed with Pγ (PDBID: 3JWR) [12], and an NMR solution structure of the intrinsically disordered rod Pγ subunit (PDBID: 2JU4) [120]. Recent structural biology studies on both rod and cone PDE6 have greatly advanced our understanding of the domain organization, subunit topology, allosteric regulation, and transducin activation mechanism.
Rod PDE6 Structure
A structural model for the rod Pαβ catalytic dimer, including novel information on the dimerization interface of Pαβ and asymmetry in the interactions of Pγ with each catalytic subunit, was reported in 2014 [143] based on high-density chemical cross-linking and LC-MS sequencing of cross-linked peptide fragments (XL-MS) in conjunction with available template structures of rod PDE6 using the Integrated Modeling Platform [134]. Subsequently, an 11 Å cryo-EM reconstruction of rod PDE6 holoenzyme complexed with antibody fragments (PDBID: 3JBQ) was reported that validated the overall domain organization of rod PDE6 and the extended conformation of Pγ that spans both of the GAFa and GAFb domains and the catalytic domain [148]. However, the reversible domain rearrangement observed by Zhang et al. [148] upon the removal and rebinding of Pγ with the Pαβ catalytic dimer has not been observed by others [70,143,114,28].
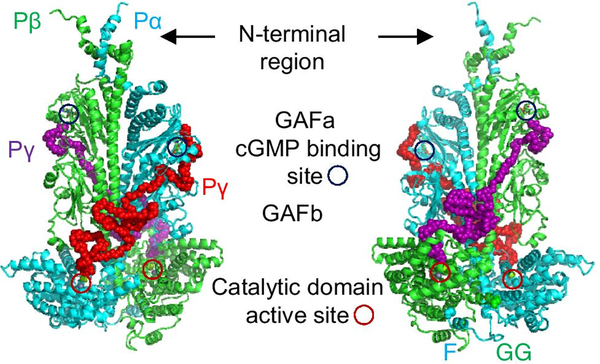
The first high-resolution (3.4 Å) cryo-EM structure of the rod PDE6 holoenzyme (PDBID: 6MZB) offered a substantial advance in our knowledge of the molecular architecture of the rod PDE6 quaternary structure [53]. Gulati et al. resolved the atomic-level structure of most of the N-terminal region of the catalytic subunits, mapped a majority of the interaction surface of each the Pγ subunit with Pαβ, and demonstrated direct allosteric communication between the GAFb domain and the catalytic domain upon binding of a PDE5/6-selective inhibitor to the enzyme active site. Using XL-MS and integrative structural modeling, Irwin et al. provided structural data for missing elements from the Gulati et al. PDE6 cryo-EM structure and elucidated the striking topological differences in the binding of each Pγ subunit to the PDE6 catalytic heterodimer (Fig. 1; [68]). This work also revealed additional elements of secondary structure in the N-terminal and C-terminal regions of Pαβ as well as provided information on flexible regions that were poorly resolved by cryo-EM (including the central region of Pγ that interacts with the GAFb and catalytic domains) [68].
Fig. 1.
Structural model of rod PDE6 holoenzyme. Rod PDE6 consists of a catalytic heterodimer (Pαβ) to which two Pγ subunits bind in an extended linear conformation. Each Pγ subunit primarily interacts with one catalytic subunit, extending from the cGMP binding GAFa domain to the GAFb domain and then crossing over to the catalytic domain where the Pγ C-terminal residues occlude the entrance to the enzyme active site in the nonactivated state. The two views, rotated by 180°, demonstrate the asymmetry of the interaction surface of each Pγ subunit with Pαβ, especially in the polycationic region of Pγ that interacts with the GAFb domain. The C-terminus of each catalytic subunit is post-translationally modified with a farnesyl group (F) or a geranylgeranyl group (GG). Adapted from [68].
Cone PDE6 Structure
The inability to obtain sufficient quantities of purified cone PDE6 holoenzyme—either from mammalian retina or by heterologous expression—for atomic-level structural studies has slowed efforts to evaluate rod-cone differences in PDE6. However, the atomic-level structure and allosteric communication pathways of the homodimeric cone GAFab regulatory domains have recently been elucidated using x-ray crystallography at 3.3 Å resolution (PDBID: 6X88) in conjunction with XL-MS experiments and integrative structural modeling to refine the solution structure by determining flexible elements that were not resolved in the crystal structure (Fig. 2) [55].
Fig. 2.
Structural model for the regulatory GAFab domains of cone PDE6 liganded with cGMP and Pγ. The two chains of the cone GAFab regulatory domains (amino acids 42–458) are colored green and dark blue, while the truncated Pγ subunits (amino acids 1 to 58) are depicted as thick orange and yellow lines. Major structural elements undergoing ligand-dependent conformational changes in the GAFa domain (red labels) and GAFb domain (orange labels) are highlighted, with arrows indicating the proposed allosteric communication pathway from GAFa to GAFb (red arrows) and from GAFb to the catalytic domain (orange arrows). Adapted from [55].
The cone GAFab structure shares the same tandem GAF domain fold as for PDE2, PDE5, and rod PDE6 [105,131,53,55], with the dimer interface extending the length of the long axis of the two central α-helices and including stabilizing contributions from an α-helix preceding the GAFa domain. The two monomeric units comprising the homodimer are highly symmetrical, with conformational differences most pronounced in the regions lacking secondary structure. The GAFab x-ray structure [55] was solved in the apo state, and a comparison with a previous crystal structure for the cGMP-bound GAFa domain [89] showed little structural differences, aside from minor movement of residues lining the cGMP binding pocket. Comparison of cone GAFab (with bound cGMP and Pγ) with the GAFab domains of rod PDE6 holoenzyme reveal the greatest structural differences in the GAFb domain (particularly the GAFb β1/β2 loop) as well as major differences in the interaction surface of Pγ with the GAF domains [55] (discussed further below). Since the Gupta et al. structure lacks the first 41 amino acids of the N-terminal region as well as the entire catalytic domain, a full understanding of the structure, conformational dynamics, and significance of the structural differences between rod and cone PDE6 must await the atomic-level structural determination of the entire cone PDE6 holoenzyme.
The next challenge is to determine the molecular dynamics of the non-identical cGMP and Pγ binding interactions with the rod and cone catalytic dimer that accompany transducin activation of PDE6 and the allosteric communication network that together regulate PDE6 activation and inactivation during rod and cone phototransduction. This will help us understand the extent to which structural and regulatory differences in rod and cone PDE6 holoenzymes account for the striking differences in the photoresponses of rod and cone photoreceptors [76,66,115], as well as enabling better predictions of the pathogenicity of human mutations in the catalytic and inhibitory subunits of PDE6 as well as possible pharmacological approaches to therapeutic interventions.
Catalytic Properties of Rod and Cone PDE6
Rod and cone PDE6 are cGMP-specific PDEs, as evidenced by in vitro measurements of the Michaelis constants for cGMP and cAMP differing by 40–60-fold (KM = ~15–20 μM for cGMP, 600–900 μM for cAMP [45,95,98]). Both rod and cone catalytic dimers, when fully activated by removal of the Pγ subunit from the holoenzyme, exhibit diffusion-controlled catalysis of cGMP (4500–5500 cGMP hydrolyzed per sec per catalytic dimer [45,95,98,114]). The catalytic domain of PDE6 consists of 16 α-helices, with the catalytic center containing tightly bound Zn2+ and Mg2+, as is the case for all eleven members of the Class I cyclic nucleotide PDEs [13]. Available evidence indicates that the active sites of Pα and Pβ have similar enzymatic properties to each other and to the cone PDE6 catalytic site [98] as well as similar binding affinity of Pα and Pβ for the potent PDE5/6 inhibitor, vardenafil [86]. Rod and cone PDE6 show no significant differences in their ability to be inhibited by a variety of PDE inhibitors [145] indicating that the structure of the catalytic pockets of all three PDE6 catalytic subunits are very similar with regards to both substrate specificity and drug binding affinity.
Allosteric Regulation of PDE6
Compared with the allosteric mechanisms responsible for regulation of other GAF-containing PDEs, the evolution of PDE6 from an ancestral PDE5/PDE6/PDE11 gene [78,80] and the co-evolution of the inhibitory Pγ subunits [78,133] provides a multifaceted level of allosteric modulation that make it uniquely suited to serve as the central effector of visual transduction. Whereas PDE2 catalytic activation occurs upon binding of cGMP to its regulatory GAFb domains resulting in increased substrate access to the active site [105], regulation of PDE6 hydrolytic activity by cGMP occupancy of the GAFa domain has not been observed. That said, inter-domain allosteric communication between the GAFa and catalytic domain of rod PDE6 has been inferred from biochemical observations ([144] and reference cited therein).
Rod PDE6 Allosteric Regulation
Occupancy of the rod PDE6 GAFa domains by cGMP has been shown to enhance the overall affinity of the Pγ subunit for Pαβ. This cooperative binding effect is reciprocal, in that binding of Pγ to Pαβ enhances cGMP binding to the two non-identical cGMP binding sites in the GAF domains [140,5,34,95]. This allosteric mechanism serves to lower the basal catalytic activity of PDE6 in its nonactivated state and may be a major factor reducing spontaneous PDE6 activation and contributing to the extraordinary light sensitivity of rod photoreceptors [117].
The inhibitory Pγ subunit is a small (~10 kDa) intrinsically disordered protein whose N-terminal half binds to the GAFab domain to enhance cGMP binding affinity [146] whereas its C-terminal half interacts with the catalytic domain to block the entrance to the catalytic site [52,12]. The Pγ subunit is a multifunctional protein (reviewed in [54]) which interacts not only with Pαβ, but also with Gtα and RGS9–1. Pγ is subject to reversible phosphorylation at two threonine residues in its polycationic region ([104] and references cited therein). Although transgenic mice in which one or both of these threonine residues were mutated to alanine residues result in alteration of the rod photoresponse [129,138] and light-induced increases in Pγ phosphorylation have been observed ex vivo [69], the significance of Pγ phosphorylation for regulation of the phototransduction pathway in vivo remains unclear.
Recent advances in the structural biology of rod and cone PDE6 have begun to shed light on the conformational changes associated with the reciprocal positive cooperativity that underlies allosteric regulation by cGMP and Pγ. The cryo-EM structure of rod PDE6 holoenzyme (with bound cGMP) provided the first structural evidence for inter-domain interactions (e.g., GAFa β1-β2 loop to GAFb domain, GAFb β1-β2 loop to catalytic domain) [53]. Chu et al. [28] used quantitative, label-free XL-MS to evaluate conformational changes in rod PDE6 in four different liganded states: apo Pαβ catalytic dimer, Pαβ with cGMP occupying the GAFa binding sites, Pαβ reconstituted with two Pγ subunits, and Pαβ with bound cGMP and two Pγ. Analysis of each liganded state identified cross-linked pairs that underwent cGMP and/or Pγ-dependent conformational changes in less structured regions of the catalytic domain (including the M-loop adjacent to the active site). Furthermore, twice as many conformationally sensitive residues were identified in Pα than Pβ (even though the Pβ subunit was more accessible to the chemical crosslinker), and Pγ cross-links with the GAFa domain were observed only for the Pα subunit [143,28]. The structural and conformational asymmetry of the Pαβ dimer complement the observations that both cGMP and Pγ exhibit two non-identical classes of binding sites with the rod heterodimer [96,95], and suggest that Pα may play the dominant role in allosteric communication between the GAF and catalytic domains.
Cone PDE6 Allosteric Regulation
Because of the challenges in obtaining purified cone PDE6 [45,64,97], much less is known about allosteric regulation of cone PDE6 by cGMP and Pγ binding to the cone catalytic homodimer.
A structural analysis of the conformational changes and allosteric regulation of cGMP and Pγ binding to cone PDE6 has recently been reported, relying on the structure of cone GAFab (described above) as a template and examining XL-MS results as well as other structural approaches to detect conformational changes resulting from cGMP binding, Pγ binding, or binding of both ligands to the GAFab regulatory domain [55]. Molecular dynamics (MD) simulations comparing the apo and cGMP-liganded cone GAFab homodimer documented conformational perturbations upon cGMP binding (consistent with a solution NMR study of the apo and cGMP-bound GAFa domain [89]), as well as structural asymmetry of the two GAFab subunits comprising the cone homodimer. Cross-linking of Pγ to GAFab in conjunction with solution NMR spectroscopy of isotopically labeled Pγ identified multiple sites within the central polycationic region of Pγ interacting with the GAFb domain [55].
From this work a set of structural elements participating in allosteric signal relay mediated by cGMP and Pγ binding to the regulatory domains of cone PDE6 has been proposed [55]. In this model, binding of cGMP to GAFa induces conformational changes in the α4 helix (which forms a “lid” to the cGMP binding pocket) and the GAFa β1/β2 loop, relaying the binding signal to the GAFb domain (Fig. 2, red highlights). Binding of Pγ and cGMP to GAFab accentuated these conformational changes, consistent with the reciprocal allosteric regulation observed between cGMP and Pγ in rod PDE6 (see above). Upon cGMP and/or Pγ binding to GAFab, the GAFb domain undergoes conformational changes most pronounced in the GAFb β1/β2, β4/β5, and β5/β6 loops as well as significant motion of the long α-helix that connects the GAFb domain to the catalytic domain (Fig. 2, orange highlights). Until studies of the conformational dynamics of the complete cone PDE6 holoenzyme can be carried out, it remains to be determined whether the allosteric communication network between the regulatory GAF domains and the catalytic domains are similar in the rod and cone PDE6 isoforms.
Physiological significance of cGMP binding to the GAFa domain of PDE6
Although the above-mentioned biochemical and structural studies have demonstrated that allosteric communication between the GAFab domain and the catalytic domain does occur in PDE6 (as is the case for the other four GAF-containing PDE families), the functional significance of cGMP binding to noncatalytic GAFa binding sites remains unclear. It has been proposed that the GAFa binding sites serve to buffer cGMP levels in the dark-adapted state and work in concert with light-activated guanylyl cyclase activity to rapidly restore cGMP levels during the recovery phase of the photoresponse [35,141]. However, the rate of cGMP dissociation from noncatalytic binding sites on rod PDE6 is much too slow [22,103] to significantly contribute to restoration of cytoplasmic cGMP levels during rod photoresponse recovery. It has also been suggested dissociation of cGMP from GAFa binding sites during prolonged illumination of rod photoreceptors that persistently lowers cytoplasmic cGMP levels would lead to decreased affinity of Pγ for Pαβ and allow Pγ to potentiate RGS9–1-catalyzed acceleration of transducin GTPase [3,34]; see below for discussion of RGS9–1 inactivation mechanism. While a role for cGMP dissociation from the noncatalytic sites during prolonged light adaptation remains plausible [41], there is currently no experimental evidence supporting this idea in rod photoreceptors [21].
A likely role for the noncatalytic cGMP binding sites in PDE6 arises from comparison of biochemical differences of rod and cone PDE6 in conjunction with the electrophysiological differences in rod and cone photoreceptors. Whereas the overall affinity of rod and cone Pγ for the rod or cone catalytic dimer are very similar [98,133], cone PDE6 holoenzyme binds cGMP to its noncatalytic sites with significantly lower affinity than rod PDE6 [45,64,95]. In combination with the reciprocal positive cooperativity between cGMP and Pγ binding mentioned above, the more rapid equilibrium of cGMP binding to the GAFa binding sites in cone PDE6 may be the mechanistic basis for cone photoreceptors exhibiting a greater level of “dark noise” (i.e., continuous fluctuations in the dark-adapted circulating current ascribed to spontaneous activation of PDE6) than rod photoreceptors [117,118], as well as exhibiting higher rates of cGMP metabolic flux [115]. From an evolutionary perspective, it is conceivable that the exquisite light sensitivity of rod photoreceptors arose from mutations in the GAFa cGMP binding sites that enhanced the affinity of cGMP for rod PDE6 and thereby reduced the frequency with which Pγ would dissociate from the catalytic dimer to cause spontaneous activation. Support for this idea comes from the observation that the number of invariant amino acids in the GAFa domain is 3-fold higher for the two rod catalytic subunits than for cone GAFa [20]; a similar phenomenon of greater sequence conservation of rod Pγ compared with cone Pγ has also been observed [133]. While an intriguing possibility, to date there has been no direct assessment of the idea that the cGMP binding sites in cone PDE6 have functional significance for regulating the basal activity of cone PDE6 or its activation/inactivation kinetics in intact cone photoreceptors. A final possibility is that cGMP binding to the GAFa domain of rod and cone PDE6 primarily serves to facilitate the proper folding of the GAFa domain during assembly of the PDE6 holoenzyme. The observation that a retinal disease-linked missense mutation in the cone PDE6 GAFa domain (Arg104Trp; [24]) leads to impaired protein stability [26] lends support for a role for cGMP binding in stabilizing the structure of PDE6.
Mechanism of PDE6 Activation by Transducin during Visual Excitation
The rod and cone photoreceptor PDE6 signaling complexes are tightly regulated during visual transduction. Under dark-adapted conditions, the high affinity of the inhibitory Pγ subunits for Pαβ is responsible for a very low rate of cGMP hydrolysis by its catalytic subunits [36]. Upon light activation of rhodopsin (Fig. 3, R*, left), the heterotrimeric G-protein (transducin, Gαβγ) undergoes nucleotide exchange and Gtα*-GTP dissociates from the transducin βγ dimer and rapidly binds to PDE6 holoenzyme to relieve the inhibitory constraint of the Pγ subunits (Fig. 3). PDE6 recovers to its nonactivated state by a deactivation mechanism that is described in detail in a later section.
Fig. 3.
Protein complexes involved in visual excitation and deactivation in rod photoreceptor cells. Binding of transducin (Gαβγ) to photoactivated rhodopsin (R*) leads to GTP exchange on Gtα and dissociation of the activated Gtα*-GTP subunit. PDE6 holoenzyme (αβγγ) is maximally activated upon binding of two Gtα*-GTP that displace the inhibitory Pγ subunits from the enzyme active site (not visible). Restoration of the nonactivated state of PDE6 involves interaction of the RGS9–1 inactivation complex with Gtα*-GTP and Pγ that accelerates the GTPase activity of Gtα*. Gtα-GDP then dissociates from PDE6, leading to the re-inhibition of PDE6 and the dissociation of Gtα-GDP to re-form the inactive transducin heterotrimer. Space-filling models were generated from the following sources: R*-Gαβγ [44]; Gtα*-PDE6 activated complex [68]; RGS9/Gβ5L [25]; R9AP homology model was generated with I-Tasser with its C-terminal domain inserted into the membrane [15].
Despite over four decades of visual transduction research, there remain major gaps in our knowledge about the mechanism by which catalytic inhibition of rod PDE6 imposed by its Pγ subunit is released upon interaction with activated Gtα*. As one example, the degree of signal amplification when a photoactivated rhodopsin (R*) catalyzes GDP/GTP exchange to sequentially activate multiple Gtα*-GTP subunits continues to be a subject of study [6,4], with recent in vivo estimates from ranging from as little as 12 Gtα* per R* to greater than a thousand Gtα* per R* (discussed recently in [79,142,81]).
Of particular relevance to this review, there also remains controversy about how activated Gtα* binds to and relieve Pγ inhibition of the PDE6 holoenzyme, including the affinity and stoichiometry of Gtα* binding to PDE6, the extent of activation (relative to the full catalytic potential of Pαβ lacking bound Pγ), and the molecular interactions at each stage of the activation process. This uncertainty can be ascribed in part to the fact that the rod PDE6 holoenzyme is a catalytic heterodimer to which two Pγ subunits bind in a functionally and structurally asymmetric manner (Fig. 1). Historically, several biochemical activation mechanisms have been postulated for rod PDE6, including: (a) binding of a single Gtα* subunit to PDE6 induces nearly full enzyme activation at both catalytic sites [18,91]; (b) binding of two Gtα* subunits per PDE6, each of which independently activates one-half of the maximum catalytic activity [137,84]; and (c) binding of two Gtα* to PDE6 occurs in a sequential manner, the first binding event causing little catalytic activation of PDE6, with the binding of the second Gtα* causing either partial [93,43] or complete [14,114] catalytic activation. The preponderance of evidence from studies that have examined the extent of PDE6 activation by Gtα* over a range of Gtα* concentrations is that Gtα* is capable of activating PDE6 holoenzyme to the same extent as Pαβ (prepared by limited trypsin proteolysis to degrade the Pγ subunits) [14,18,91,86,147,68]. The consensus from both biochemical [137,29,114] and structural studies [68] support the requirement for two Gtα* molecules to bind to PDE6 to achieve full activation. The prevailing view is that the two Gtα* binding sites on PDE6 exhibit large differences in binding affinity [14,93,114].
Our knowledge of the activation mechanism of cone PDE6 is mainly derived from transgenic mice studies in which rod and cone homologs of opsin, transducin, or PDE6 have been manipulated (reviewed in [66]). In the absence of obvious differences in the biochemical properties of rod and cone opsin, transducin, or PDE6 catalytic subunits, Wang et al. [133] conducted a phylogenetic analysis of rod and cone Pγ sequences and used this information to evaluate whether sequence differences of rod and cone Pγ might account for the greater in vitro effectiveness of cone PDE6 to become activated by rod transducin than rod PDE6 [45]. Wang et al. discovered that a rod Pγ-specific sequence motif (amino acids 9–18) suppressed efficient GTα* activation of PDE6 whereas two cone Pγ-specific amino acid residues (Asn-13 and Gln-14) facilitated transducin activation of PDE6. This work identified a novel role for the N-terminal region of Pγ for regulating the extent of Gtα* activation of PDE6 in vitro, adding yet another element of regulatory significance to this multifunctional, intrinsically disordered inhibitory subunit of PDE6 [54]. Future work is needed to reconcile these in vitro observations on rod-cone differences in the Pγ subunit with well-established physiological differences in the light sensitivity and kinetics of rod and cone photoresponses [66]; indeed, it would be interesting to ascertain the consequences of using transgenic mice in which rod Pγ was substituted for cone Pγ in rod photoreceptors, as was done with the cone catalytic subunit [87].
Molecular Architecture of the Gtα*-PDE6 Activation Complex
Two different reports of the topology of the Gtα*-PDE6 activated complex have come to somewhat different conclusions about the mechanism of Gtα* binding to the PDE6 holoenzyme and relieving Pγ inhibition of catalysis. Irwin et al. conducted XL-MS experiments of the complex of an aluminum fluoride-activated transducin α-subunit (Gtα*-GDP-AlF, in which AlF4−acts as an analog of the terminal phosphate of GTP [16]) and PDE6 holoenzyme (both isolated from bovine rod outer segments) reconstituted on liposomal membranes, as well as reporting on the Pγ-Gtα* complex [68]. It was first observed that tethering Gtα to the membrane causes the N-terminal α-helix (αN) to come into proximity to the guanine nucleotide binding site of Gtα, in contrast to the conformation of the N-terminus observed in the crystal structure of the transducin heterotrimer (PDBID: 1G0T [82]). Upon addition of Pγ to Gtα*-GDP-AlF, several cross-links were detected between the polycationic region of Pγ (residues 25, 39, and 45) and the Gtα switch II region and the αB and αC elements of the helical domain [68], consistent with earlier biochemical studies [8,147]. Upon formation of the membrane-confined Gtα*-GDP-AlF-PDE6 activated complex (that activated PDE6 catalysis to the same extent as Pαβ), Irwin et al. characterized two different sets of Gtα* docking sites from their crosslinking data (Fig. 4A and 4B): (a) numerous crosslinks defined an interface of interaction of two Gtα* molecules with the Pα and Pβ catalytic domains (Fig. 4A), consistent with previous mutagenesis studies [101,51] and analogous to the configuration of the complex of adenylyl cyclase with Gsα [113]; (b) Pα- and Pβ-specific crosslinks between the GAFb domains and Gtα* generated structural models (Fig. 4B) implicating docking of Gtα to the polycationic region of Pγ that is localized to the GAFb domains of nonactivated PDE6 [68]. Although the conformational changes of Pγ upon transducin activation of PDE6 could not be resolved in this study, the overall results are suggestive of the following sequential activation mechanism: (a) the first Gtα* molecule initially binds to the GAFb domain of nonactivated PDE6 holoenzyme (Fig. 4B) without significantly inducing PDE6 activation, due to high affinity interactions of Gtα* with the Pγ polycationic region (see Fig. 1); (b) binding of Gtα* to Pγ weakens Pγ interactions with the PDE6 GAFb domain, causing Gtα* (complexed with the polycationic region of Pγ) to migrate to the catalytic domain (Fig. 4A), and also inducing the N-terminal portion of Pγ to be displaced from its interactions with GAFa (consistent with biochemical studies of elevated rates of cGMP dissociation from GAFa upon Gtα* activation of PDE6 [103,146]; (c) these structural rearrangements facilitate binding of the second Gtα* to the other catalytic subunit, resulting in displacement of both Pγ C-terminal regions from the enzyme active sites and full catalytic activation of PDE6 (see Fig. 4 of [68] for details).
Fig. 4.
Comparison of structural models for the proposed interactions of PDE6 with activated Gtα* a. Structural model for Gtα* docking to the catalytic domain of membrane-associated rod PDE6 (adapted from Fig. 3A of {Irwin, 2019 #8185}). b. Model of Gtα* binding to the GAFb domain of PDE6 (from Fig. 3C of {Irwin, 2019 #8185}); c. Structural model of chimeric Gtα/Giα docking to GAFb region of rod PDE6 (adapted from {Gao, 2020 #8312}; PDBID: 7jsn). In each panel, the three structures were aligned to the PDE6 catalytic dimer (Pα, cyan; Pβ, green). Only one of the two G-protein subunits bound to PDE6 is shown for the sake of clarity with α-helical elements colored to visualize conformational differences of the binding interfaces (N-terminal αN helix, magenta; αC and αD helices within the helical domain, yellow; α3 region of Ras domain, blue; C-terminal α5 helix, red).
Gao et al. subsequently reported the 3.2 Å cryo-EM structure of a soluble complex of a transducin/Giα chimera and PDE6 holoenzyme (PDBID: 7jsn) in which the PDE5/6 inhibitor vardenafil occupies the PDE6 catalytic sites and the activated complex was formed by use of a bivalent antibody that tethered two Gtα* molecules to stabilize the Gtα*-PDE6 complex [43]. The two Gtα* subunits are docked to the GAFb domain of Pα and Pβ (Fig. 4C) in an upside-down configuration (relative to the presumed Gtα* orientation when associated with the membrane) and form numerous interactions with the Pγ subunits whose C-terminal region has been completely displaced from its association with the catalytic domain without significant changes in the interactions of the N-terminal region of Pγ with the GAFa domains [43]. Evaluation of the dynamic elements of the Gtα*-PDE6 complex by multi-body refinement and principal component analysis revealed concerted motions of the two Gtα* subunits consistent with an alternating-site catalytic mechanism that involves inter-subunit communication between Pα and Pβ as well as fluctuations in Pγ conformation; this mechanism would relieve Pγ inhibition at only one active site at a time, and hence lead to a maximum of 50% of the activity of Pαβ lacking Pγ) [43].
The differences in the structural models presented by these two groups highlight limitations in each study. While the advantages of the XL-MS study include assembly of the Gtα*-PDE6 complex on the membrane with native, purified proteins in a more physiological orientation, and the ability to sample an ensemble of conformations, a major drawback is the relatively low resolution of this approach including the need to identify multiple crosslinks to generate sufficient distance constraints for accurate modeling. Although the cryo-EM study offers atomic-level resolution of the Gtα*-PDE6 complex, the use of a bivalent antibody to bind two Gtα* molecules in order to obtain stable protein complexes with PDE6 may have prevented Gtα* docking to the PDE6 holoenzyme in a manner that occurs in vivo; this concern is heightened by the fact that occupancy of the active site by vardenafil (to stabilize the Gtα*-PDE6 complex for cryo-EM) is likely to have weakened Pγ affinity for the catalytic domain due to competition between the drug and Pγ for the active site [145]. Future advances in determining the mechanism of transducin activation of PDE6 will require investigating the sequence of binding events and the conformational dynamics that occur when each of the two Gtα* subunits interact with a catalytic subunit and its bound Pγ to release the catalytic potential of the PDE6 active sites during visual excitation.
Inactivation of PDE6 by RGS9–1 during Recovery to the Dark-Adapted State
Under most physiological conditions, the rate-limiting step for recovery of the photoresponse in rod and cone photoreceptors is the inactivation of the Gtα*-PDE activated state that is regulated by the GTPase accelerating protein (GAP) RGS9–1 and its binding partners (Fig. 3) [77,7,107]. RGS9–1 is a photoreceptor-specific splice variant of the R7 sub-family of RGS proteins [58,2]. The RGS9–1 domain organization consists of an RGS catalytic domain (responsible for its GAP activity), a G protein γ-like (GGL) domain that interacts with the Gβ5L protein (a member of the G protein β-subunit family), and the N-terminal DEP (Dishevelled, Egl10, Pleckstrin) and DEP helical extension (DHEX) domains. The DEP domain is believed to interact with the RGS9–1 anchoring protein R9AP [63,85]. The PDE6 Pγ subunit is known to potentiate the intrinsic GAP activity of RGS9–1 [3], with enhancement of GAP activity occurring when Gtα*-GTP is reconstituted with the entire RGS9–1/Gβ5 heterodimer [59]. Binding of R9AP to RGS9–1/Gβ5 is reported to further enhance GAP activity by a distinct mechanism [85,88,11]. In addition, reversible phosphorylation of RGS9–1 near its C-terminus and other light-dependent reactions have been suggested to influence R9AP binding affinity and/or potentiation of GAP activity ([125] and references cited therein).
Whereas the biochemical mechanism of RGS9–1-catalyzed acceleration of transducin GTPase activity and the physiological role of RGS9–1 as the rate-limited step in the recovery of the photoresponse have been well studied, the molecular organization of the PDE6 deactivation complex and the sequence of steps by which RGS9–1 binds to the activated Gtα*-PDE6 complex, engages the Pγ subunit to facilitate acceleration of GTPase activity, and then dissociates to restore the nonactivated state of transducin and PDE6 remains to be explored.
Tissue and Subcellular Distribution of PDE6
The tissue distribution of PDE6 is quite restricted compared with other PDE family members. Rod PDE6 holoenzyme is localized to the disk membranes of photoreceptor outer segments at high concentrations (80 PDE6 holoenzyme molecules per μm2 of disk membrane surface); this represents a PDE6:transducin:rhodopsin ratio of 1:30:300 [108]; the density of cone phototransduction proteins on the corresponding membrane surface of cone outer segments is less well understood. In addition, the mammalian pineal gland has been shown to contain not only PDE6 but also other phototransduction proteins [23,17,62]. While there are reports of PDE6 catalytic or inhibitory subunits being expressed in non-photoreceptive tissues or in tumor cells [123,102,40,48,150], the functional significance is uncertain because in many instances the work is based only on the presence of mRNA transcripts without verification of protein expression. Furthermore, co-expression of PDE6 catalytic and inhibitory subunits outside of photoreceptive tissues has not yet been reported, raising questions about the functional significance of individual PDE6 catalytic genes being expressed or of novel binding partners for the Pγ subunit in the absence of PDE6 catalytic subunits.
PDE6 Pharmacology
PDE Inhibitor Compounds
Drugs targeting cyclic nucleotide PDEs have largely focused on compounds intended to bind to the enzyme active site with high affinity and thereby block the catalysis of cyclic nucleotides [90]. This therapeutic strategy has seen remarkable success with the introduction of numerous FDA-approved PDE inhibitors that selectively target individual PDE families, and, in a few instances, specific isoforms of a particular PDE family [9]. Since there are no obvious clinical applications for inhibitor compounds targeting rod and cone PDE6, most pharmacological research on PDE6 has focused on designing inhibitor compounds for other PDE families that have low affinity for PDE6 [32].
PDE6 is most closely related to the PDE5 enzyme family, based on primary sequence, three-dimensional structure, substrate specificity, as well as known susceptibility to inhibition by compounds designed to target the PDE5 active site [32]. Whereas most so-called PDE5-selective inhibitors are more accurately characterized as PDE5/6-selective inhibitors due to lack of selectivity for PDE5 over rod or cone PDE6 (e.g., sildenafil, vardenafil [145]), a few compounds have been identified that inhibit PDE5 with 100-fold or greater potency than PDE6 (e.g., tadalafil [39,145]). PDE5/6 inhibitors paradoxically elevate the hydrolytic activity of nonactivated PDE6, which has been shown to result from the competition between the Pγ subunit, the PDE5/6 inhibitor, and substrate for binding to the active site [46,145]. Mutagenesis and structural studies have provided insights into the differences in the PDE5 and PDE6 drug binding sites [20,106,43] that will aid future structure-guided drug design of next-generation PDE5 inhibitors with greater selectivity for PDE5 over PDE6 [65,72].
Overall, administration of PDE5-targeted inhibitors has not been correlated with major adverse effects on retinal function in healthy individuals, but their use is contraindicated for individuals diagnosed with hereditary retinal disease [83]. Minor adverse side effects to visual function associated with administration of approved PDE5 inhibitors can arise from direct effects on inhibition of PDE6 in rod and cone photoreceptors (e.g., impaired color vision, photophobia) as well as by inhibition of PDE5 that is present in vascular smooth muscle of ocular blood vessels [74,9,37].
Given the evidence that elevated cGMP levels in retinal photoreceptors are cytotoxic and lead to photoreceptor cell death (see next section), pharmacological approaches to elevating PDE6 activity hold promise for neuroprotection of photoreceptors. One potential drug target is the cGMP-binding GAFa domains of rod and cone PDE6; compounds that displace cGMP from its GAFa binding sites might be expected to elevate the basal activity of PDE6, given the positive cooperativity between cGMP binding and Pγ binding affinity (see section on Allosteric Regulation of PDE6). Another possible strategy to modulate PDE6 activity would be to identify small molecules or interfering peptides [10,19] that could serve as allosteric modulators of catalytic activity or alternatively alter Pγ interactions with PDE6 catalytic subunits (e.g., short Pγ peptides that compete with endogenous Pγ and enhance hydrolytic activity [95]). The ability to counteract the cytotoxic effects of excessive cGMP accumulation by pharmacologically manipulating the activity of PDE6 would have applicability to a number of inherited retinal diseases, making this approach an attractive alternative to gene therapy that targets a single defective gene [124].
Molecular Etiology of Retinal Diseases Resulting from Mutations in PDE6
Some inherited retinal diseases are caused by dysregulation of the phototransduction pathway in rod and cone photoreceptors that lead to an excessive accumulation of cGMP. This can result in an influx of calcium ions and/or activation of protein kinase G and, ultimately, photoreceptor cell death [132,49,67,126] (see accompanying article by Francois Paquet-Durand). These retinal diseases include retinitis pigmentosa [56,38], cone-rod dystrophy [57,128], congenital stationary night blindness [127], Leber congenital amaurosis [75] and achromatopsia [116,61]. Impaired regulation of photoresponse recovery or of light adaptation arising from prolonged activation of the phototransduction pathway can also lead to visual disorders (e.g., bradyopsia [92,121]).
Mutations in the genes coding for rod and cone PDE6 (PDE6A, PDE6B, PDE6G, and PDE6H) account for a significant fraction of the retinal degenerative diseases (cataloged at the Retinal Information Network: https://sph.uth.edu/RETNET/). Indeed, the identification of a naturally occurring mutation in PDE6B gene in the rd1 mouse was perhaps the earliest identification of a genetic defect associated with retinal degeneration [73,119,109]. Since then, substantial progress has been made in understanding the physiological consequences of mutations in PDE6 genes through the use of transgenic mice in which a PDE6 gene [or the PDE6 chaperone, AIPL1 [50]] has been mutated (reviewed in [130,30]). Dysregulation of cGMP metabolism due to these inherited defects in structure, function and/or regulation of PDE6 typically results in complex phenotypes leading to photoreceptor cell death (for review, see [111]).
In addition to in vivo characterization of selected PDE6 mutations using animal models, advances over the past decade in heterologous expression of PDE6 catalytic subunits—either through ectopic expression of constructs in transgenic X. laevis photoreceptors [99,98,27,26] or, more recently, through heterologous co-expression of PDE6 catalytic and Pγ subunits with AIPL1 in mammalian cell culture [50,49]—have enabled the study of the molecular etiology of PDE6 mutations underlying retinal degenerative diseases. The combination of in vivo and heterologous expression studies permits the categorization of disease-causing PDE6 mutations into several (not mutually exclusive) categories: (1) aberrant folding and/or subunit assembly of PDE6 catalytic and inhibitory subunits with AIPL1 during biosynthesis in the inner segment leading to disturbances in the protein homeostasis network; (2) abnormal trafficking of PDE6 from the inner segment to its destination on the outer segment membrane; (3) loss of catalytic activity due to alterations in the conformation of the enzyme active site; (4) elevated catalytic activity arising from an inability of Pγ to occlude the active site in its nonactivated state; (5) disruption of the allosteric communication network between the GAF domains and the catalytic domain; and (6) alterations in the surface of interaction of PDE6 with PDE6-interacting proteins (e.g., transducin, RGS9–1, protein kinases).
However, we still lack a mechanistic understanding of the structure, conformational dynamics, and regulation of PDE6 and its binding partners needed for the rational design of novel therapeutic interventions for retinal diseases (see previous section). Recent developments in predicting the pathogenicity of human mutations have emphasized the importance of understanding not just the amino acid sequence and 3-dimensional structure (e.g., PolyPhen-2 [1]) but also the conformational dynamics and binding interfaces of mutated proteins [110]. The importance of understanding the structure, protein dynamics, and molecular organization of PDE6 interacting proteins is highlighted by the following observations: (1) several conformationally sensitive residues of rod Pαβ are in proximity to known human PDE6 missense mutations [28]; (2) a number of disease-causing mutations in the cone PDE6 GAFab domain are localized to sites that undergo ligand-dependent conformational changes, including the GAFa β1/β2 loop and the GAFb β1/β2 loop (see Fig. 8 of [55]); (3) a disease-causing missense mutation in the Gtα subunit (Q200E [122]) is located in the interface of interaction of Gtα with rod PDE6 [68,43] while another Gtα mutation (D129G [100] is within 10 Å of the polycationic region of Pγ that binds to Gtα [68]). The prevalence of disease-causing mutations in flexible, conformationally dynamic regions of PDE6 subunits support the hypothesis that therapeutic strategies targeting the allosteric communication pathway of PDE6 and its interface of interaction with binding partners offers promise for pharmacological interventions designed to ameliorate dysregulation of cGMP metabolism and enhance photoreceptor health and survival.
Acknowledgements:
The author is most grateful for the scientific contributions of past and present members of his laboratory, as well as collaborating investigators. The work in the author’s laboratory is supported by the National Eye Institute (R01 EY005798) and the National Institute of General Medical Sciences (P20 GM113131) of the National Institutes of Health.
Funding: The research in the author’s laboratory was supported by the National Eye Institute (NIH) grant R01 EY05798, and the National Institute of General Medical Sciences (NIH) grant P20 GM113131.
Footnotes
Conflicts of interest/competing interests: The author has no relevant financial or proprietary interests in any material discussed in this article.
Declarations
Ethics approval: Not applicable.
Consent to participate: Not applicable.
Consent for publication: Note applicable.
Availability of data and material: Not applicable.
Code availability: Not applicable.
Reference List
- 1.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson GR, Posokhova E, Martemyanov KA (2009) The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys 54:33–46. doi: 10.1007/s12013-009-9052-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arshavsky VY, Bownds MD (1992) Regulation of deactivation of photoreceptor G protein by its target enzyme and cGMP. Nature 357:416–417. doi: 10.1038/357416a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arshavsky VY, Burns ME (2014) Current understanding of signal amplification in phototransduction. Cellular Logistics 4:e29390. doi: 10.4161/cl.29390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arshavsky VY, Dumke CL, Bownds MD (1992) Noncatalytic cGMP binding sites of amphibian rod cGMP phosphodiesterase control interaction with its inhibitory γ-subunits. A putative regulatory mechanism of the rod photoresponse. J Biol Chem 267:24501–24507. [PubMed] [Google Scholar]
- 6.Arshavsky VY, Lamb TD, Pugh EN (2002) G proteins and phototransduction. Annual Review of Physiology 64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229 [DOI] [PubMed] [Google Scholar]
- 7.Arshavsky VY, Wensel TG (2013) Timing Is Everything: GTPase Regulation in Phototransduction. Investigative Ophthalmology & Visual Science 54:7725–7733. doi: 10.1167/iovs.13-13281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artemyev NO, Mills JS, Thornburg KR, Knapp DR, Schey KL, Hamm HE (1993) A site on transducin α−subunit of interaction with the polycationic region of cGMP phosphodiesterase inhibitory subunit. J Biol Chem 268:23611–23615. [PubMed] [Google Scholar]
- 9.Baillie GS, Tejeda GS, Kelly MP (2019) Therapeutic targeting of 3’,5’-cyclic nucleotide phosphodiesterases: inhibition and beyond. Nat Rev Drug Discov 18:770–796. doi: 10.1038/s41573-019-0033-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakail M, Ochsenbein F (2016) Targeting protein–protein interactions, a wide open field for drug design. Comptes Rendus Chimie 19:19–27. doi: 10.1016/j.crci.2015.12.004 [DOI] [Google Scholar]
- 11.Baker SA, Martemyanov KA, Shavkunov AS, Arshavsky VY (2006) Kinetic mechanism of RGS9–1 potentiation by R9AP. Biochemistry 45:10690–10697. doi: 10.1021/bi060376a [DOI] [PubMed] [Google Scholar]
- 12.Barren B, Gakhar L, Muradov H, Boyd KK, Ramaswamy S, Artemyev NO (2009) Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. EMBO J 28:3613–3622. doi: 10.1038/emboj.2009.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beavo JA, Francis SH, Houslay MD (2006) Cyclic Nucleotide Phosphodiesterases in Health and Disease. CRC Press, Boca Raton, FL. [Google Scholar]
- 14.Bennett N, Clerc A (1989) Activation of cGMP phosphodiesterase in retinal rods: mechanism of interaction with the GTP-binding protein (transducin). Biochemistry 28:7418–7424. doi: 10.1021/bi00444a040 [DOI] [PubMed] [Google Scholar]
- 15.Bernier SC, Horchani H, Salesse C (2015) Structure and Binding of the C-Terminal Segment of R9AP to Lipid Monolayers. Langmuir 31:1967–1979. doi: 10.1021/la503867h [DOI] [PubMed] [Google Scholar]
- 16.Bigay J, Deterre P, Pfister C, Chabre M (1987) Fluoride complexes of aluminium or beryllium act on G-proteins as reversibly bound analogues of the γ phosphate of GTP. EMBO J 6:2907–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackshaw S, Snyder SH (1997) Developmental expression pattern of phototransduction components in mammalian pineal implies a light-sensing function. Journal of Neuroscience 17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruckert F, Catty P, Deterre P, Pfister C (1994) Activation of phosphodiesterase by transducin in bovine rod outer segments: Characteristics of the successive binding of two transducins. Biochemistry 33:12625–12634. doi: 10.1021/bi00208a013 [DOI] [PubMed] [Google Scholar]
- 19.Bruzzoni-Giovanelli H, Alezra V, Wolff N, Dong CZ, Tuffery P, Rebollo A (2018) Interfering peptides targeting protein-protein interactions: the next generation of drugs? Drug Discov Today 23:272–285. doi: 10.1016/j.drudis.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 20.Cahill KB, Quade JH, Carleton KL, Cote RH (2012) Identification of amino acid residues responsible for the selectivity of tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6. J Biol Chem 287:41406–41416. doi: 10.1074/jbc.M112.389189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvert PD, Govardovskii VI, Arshavsky VY, Makino CL (2002) Two temporal phases of light adaptation in retinal rods. Journal of General Physiology 119:129–146. doi: 10.1085/jgp.119.2.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvert PD, Ho TW, LeFebvre YM, Arshavsky VY (1998) Onset of feedback reactions underlying vertebrate rod photoreceptor light adaptation. Journal of General Physiology 111:39–51. doi: 10.1085/jgp.111.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carcamo B, Hurwitz MY, Craft CM, Hurwitz RL (1995) The mammalian pineal expresses the cone but not the rod cyclic GMP phosphodiesterase. J Neurochem 65:1085–1092. doi: 10.1046/j.1471-4159.1995.65031085.x. [DOI] [PubMed] [Google Scholar]
- 24.Chang B, Grau T, Dangel S, Hurd R, Jurklies B, Sener EC, Andreasson S, Dollfus H, Baumann B, Bolz S, Artemyev N, Kohl S, Heckenlively J, Wissinger B (2009) A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci U S A 106:19581–19586. doi: 10.1073/pnas.0907720106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J (2008) Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat Struct Mol Biol 15:155–162. doi: 10.1038/nsmb.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheguru P, Majumder A, Artemyev NO (2015) Distinct patterns of compartmentalization and proteolytic stability of PDE6C mutants linked to achromatopsia. Mol Cell Neurosci 64:1–8. doi: 10.1016/j.mcn.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheguru P, Zhang Z, Artemyev NO (2014) The GAFa domain of phosphodiesterase-6 contains a rod outer segment localization signal. Journal of Neurochemistry 129:256–263. doi: 10.1111/jnc.12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu F, Hogan D, Gupta R, Gao XZ, Nguyen HT, Cote RH (2019) Allosteric regulation of rod photoreceptor phosphodiesterase 6 (PDE6) elucidated by chemical cross-linking and quantitative mass spectrometry. J Mol Biol 243:3677–3689. doi: 10.1016/j.jmb.2019.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clerc A, Bennett N (1992) Activated cGMP phosphodiesterase of retinal rods. A complex with transducin α subunit. J Biol Chem 267:6620–6627. [PubMed] [Google Scholar]
- 30.Collin GB, Gogna N, Chang B, Damkham N, Pinkney J, Hyde LF, Stone L, Naggert JK, Nishina PM, Krebs MP (2020) Mouse Models of Inherited Retinal Degeneration with Photoreceptor Cell Loss. Cells 9. doi: 10.3390/cells9040931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti M, Beavo JA (2007) Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem 76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444 [DOI] [PubMed] [Google Scholar]
- 32.Cote RH (2004) Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int J Impot Res 16:S28–S33. doi: 10.1038/sj.ijir.3901212 [DOI] [PubMed] [Google Scholar]
- 33.Cote RH (2006) Photoreceptor phosphodiesterase (PDE6): a G-protein-activated PDE regulating visual excitation in rod and cone photoreceptor cells. In: Beavo JA, Francis SH, Houslay MD (eds) Cyclic Nucleotide Phosphodiesterases in Health and Disease. CRC Press, Boca Raton, FL, pp 165–193. [Google Scholar]
- 34.Cote RH, Bownds MD, Arshavsky VY (1994) cGMP binding sites on photoreceptor phosphodiesterase: Role in feedback regulation of visual transduction. Proc Natl Acad Sci U S A 91:4845–4849. doi: 10.1073/pnas.91.11.4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cote RH, Brunnock MA (1993) Intracellular cGMP concentration in rod photoreceptors is regulated by binding to high and moderate affinity cGMP binding sites. J Biol Chem 268:17190–17198. [PubMed] [Google Scholar]
- 36.D’Amours MR, Cote RH (1999) Regulation of photoreceptor phosphodiesterase catalysis by its noncatalytic cGMP binding sites. Biochem J 340:863–869. [PMC free article] [PubMed] [Google Scholar]
- 37.da Cruz NFS, Polizelli MU, Cezar LM, Cardoso EB, Penha F, Farah ME, Rodrigues EB, Novais EA (2020) Effects of phosphodiesterase type 5 inhibitors on choroid and ocular vasculature: a literature review. Int J Retina Vitreous 6:38. doi: 10.1186/s40942-020-00241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daiger SP, Sullivan LS, Bowne SJ (2013) Genes and mutations causing retinitis pigmentosa. Clin Genet 84:132–141. doi: 10.1111/cge.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daugan A, Grondin P, Ruault C, Le Monnier de Gouville AC, Coste H, Linget JM, Kirilovsky J, Hyafil F, Labaudiniere R (2003) The discovery of tadalafil: a novel and highly selective PDE5 inhibitor. 2: 2,3,6,7,12,12a-hexahydropyrazino[1’,2’:1,6]pyrido[3,4-b]indole-1,4-dione analogues. J Med Chem 46:4533–4542. doi: 10.1021/jm0300577 [DOI] [PubMed] [Google Scholar]
- 40.Dong H, Claffey KP, Brocke S, Epstein PM (2013) Expression of phosphodiesterase 6 (PDE6) in human breast cancer cells. Springerplus 2:680. doi: 10.1186/2193-1801-2-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fain GL (2011) Adaptation of mammalian photoreceptors to background light: putative role for direct modulation of phosphodiesterase. Mol Neurobiol 44:374–382. doi: 10.1007/s12035-011-8205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francis SH, Blount MA, Corbin JD (2011) Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev 91:651–690. doi: 10.1152/physrev.00030.2010 [DOI] [PubMed] [Google Scholar]
- 43.Gao Y, Eskici G, Ramachandran S, Poitevin F, Seven AB, Panova O, Skiniotis G, Cerione RA (2020) Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6. Mol Cell 80:237–245 e234. doi: 10.1016/j.molcel.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y, Hu H, Ramachandran S, Erickson JW, Cerione RA, Skiniotis G (2019) Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation. Mol Cell 75:781–790 e783. doi: 10.1016/j.molcel.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillespie PG, Beavo JA (1988) Characterization of a bovine cone photoreceptor phosphodiesterase purified by cyclic GMP-Sepharose chromatography. J Biol Chem 263:8133–8141. [PubMed] [Google Scholar]
- 46.Gillespie PG, Beavo JA (1989) Inhibition and stimulation of photoreceptor phosphodiesterases by dipyridamole and M&B 22,948. Mol Pharmacol 36:773–781. [PubMed] [Google Scholar]
- 47.Goc A, Chami M, Lodowski DT, Bosshart P, Moiseenkova-Bell V, Baehr W, Engel A, Palczewski K (2010) Structural characterization of the rod cGMP phosphodiesterase 6. Journal of Molecular Biology 401:363–373. doi: 10.1016/j.jmb.2010.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golovastova MO, Bazhin AV, Philippov PP (2014) Cancer-retina antigens -- a new group of tumor antigens. Biochemistry (Mosc) 79:733–739. doi: 10.1134/S000629791408001X [DOI] [PubMed] [Google Scholar]
- 49.Gopalakrishna KN, Boyd K, Artemyev NO (2017) Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell Signal 37:74–80. doi: 10.1016/j.cellsig.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gopalakrishna KN, Boyd K, Yadav RP, Artemyev NO (2016) Aryl hydrocarbon receptor-interacting protein-like 1 is an obligate chaperone of phosphodiesterase 6 and is assisted by the gamma-subunit of its client. J Biol Chem 291:16282–16291. doi: 10.1074/jbc.M116.737593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Granovsky AE, Artemyev NO (2001) A conformational switch in the inhibitory γ-subunit of PDE6 upon enzyme activation by transducin. Biochemistry 40:13209–13215. doi: 10.1021/bi011127j [DOI] [PubMed] [Google Scholar]
- 52.Granovsky AE, Natochin M, Artemyev NO (1997) The γ subunit of rod cGMP-phosphodiesterase blocks the enzyme catalytic site. J Biol Chem 272:11686–11689. doi: 10.1074/jbc.272.18.11686 [DOI] [PubMed] [Google Scholar]
- 53.Gulati S, Palczewski K, Engel A, Stahlberg H, Kovacik L (2019) Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases. Sci Adv 5:eaav4322. doi: 10.1126/sciadv.aav4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo LW, Ruoho AE (2008) The retinal cGMP phosphodiesterase γ-subunit - a chameleon. Curr Protein Pept Sci 9:611–625. doi: 10.2174/138920308786733930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta R, Liu Y, Wang H, Nordyke CT, Puterbaugh RZ, Cui W, Varga K, Chu F, Ke H, Vashisth H, Cote RH (2020) Structural analysis of the regulatory GAF domains of cGMP phosphodiesterase elucidates the allosteric communication pathway. J Mol Biol 432:5765–5783. doi: 10.1016/j.jmb.2020.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamel C (2006) Retinitis pigmentosa. Orphanet J Rare Dis 1:40. doi: 10.1186/1750-1172-1-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamel CP (2007) Cone rod dystrophies. Orphanet J Rare Dis 2:7. doi: 10.1186/1750-1172-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He W, Cowan CW, Wensel TG (1998) RGS9, a GTPase accelerator for phototransduction. Neuron 20:95–102. doi: 10.1016/s0896-6273(00)80437-7 [DOI] [PubMed] [Google Scholar]
- 59.He W, Lu L, Zhang X, El-Hodiri HM, Chen CK, Slep KC, Simon MI, Jamrich M, Wensel TG (2000) Modules in the photoreceptor RGS9–1-Gβ5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J Biol Chem 275:37093–37100. doi: 10.1074/jbc.M006982200 [DOI] [PubMed] [Google Scholar]
- 60.Heikaus CC, Pandit J, Klevit RE (2009) Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights. Structure 17:1551–1557. doi: 10.1016/j.str.2009.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirji N, Aboshiha J, Georgiou M, Bainbridge J, Michaelides M (2018) Achromatopsia: clinical features, molecular genetics, animal models and therapeutic options. Ophthalmic Genet 39:149–157. doi: 10.1080/13816810.2017.1418389 [DOI] [PubMed] [Google Scholar]
- 62.Holthues H, Vollrath L (2004) The phototransduction cascade in the isolated chick pineal gland revisited. Brain Res 999:175–180. doi: 10.1016/j.brainres.2003.11.059 [DOI] [PubMed] [Google Scholar]
- 63.Hu G, Wensel TG (2002) R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9–1. Proc Natl Acad Sci U S A 99:9755–9760. doi: 10.1073/pnas.152094799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang D, Hinds TR, Martinez SE, Doneanu C, Beavo JA (2004) Molecular determinants of cGMP-binding to chicken cone photoreceptor phosphodiesterase. J Biol Chem 279:48143–48151. doi: 10.1074/jbc.M404338200 [DOI] [PubMed] [Google Scholar]
- 65.Huang YY, Li Z, Cai YH, Feng LJ, Wu Y, Li X, Luo HB (2013) The molecular basis for the selectivity of tadalafil toward phosphodiesterase 5 and 6: a modeling study. J Chem Inf Model 53:3044–3053. doi: 10.1021/ci400458z [DOI] [PubMed] [Google Scholar]
- 66.Ingram NT, Sampath AP, Fain GL (2016) Why are rods more sensitive than cones? J Physiol 594:5415–5426. doi: 10.1113/JP272556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iribarne M, Masai I (2018) Do cGMP levels drive the speed of photoreceptor degeneration? Adv Exp Med Biol 1074:327–333. doi: 10.1007/978-3-319-75402-4_40 [DOI] [PubMed] [Google Scholar]
- 68.Irwin MJ, Gupta R, Gao XZ, Cahill KB, Chu F, Cote RH (2019) The molecular architecture of photoreceptor phosphodiesterase 6 (PDE6) with activated G protein elucidates the mechanism of visual excitation. J Biol Chem 294:19486–19497. doi: 10.1074/jbc.RA119.011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janisch KM, Kasanuki JM, Naumann MC, Davis RJ, Lin CS, Semple-Rowland S, Tsang SH (2009) Light-dependent phosphorylation of the gamma subunit of cGMP-phophodiesterase (PDE6gamma) at residue threonine 22 in intact photoreceptor neurons. Biochem Biophys Res Commun 390:1149–1153. doi: 10.1016/j.bbrc.2009.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kajimura N, Yamazaki M, Morikawa K, Yamazaki A, Mayanagi K (2002) Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J Struct Biol 139:27–38. doi: 10.1016/S1047-8477(02)00502-6 [DOI] [PubMed] [Google Scholar]
- 71.Kameni Tcheudji JF, Lebeau L, Virmaux N, Maftei CG, Cote RH, Lugnier C, Schultz P (2001) Molecular organization of bovine rod cGMP-phosphodiesterase 6. J Mol Biol 310:781–791. doi: 10.1006/jmbi.2001.4813 [DOI] [PubMed] [Google Scholar]
- 72.Kayık G, Tüzün NŞ, Durdagi S (2017) Investigation of PDE5/PDE6 and PDE5/PDE11 selective potent tadalafil-like PDE5 inhibitors using combination of molecular modeling approaches, molecular fingerprint-based virtual screening protocols and structure-based pharmacophore development. Journal of Enzyme Inhibition and Medicinal Chemistry 32:311–330. doi: 10.1080/14756366.2016.1250756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keeler CE (1924) The inheritance of a retinal abnormality in white mice. PNAS 10:329–333. doi: 10.1073/pnas.10.7.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kerr NM, Danesh-Meyer HV (2009) Phosphodiesterase inhibitors and the eye. Clin Exp Ophthalmol 37:514–523. doi: 10.1111/j.1442-9071.2009.02070.x [DOI] [PubMed] [Google Scholar]
- 75.Kondkar AA, Abu-Amero KK (2019) Leber congenital amaurosis: Current genetic basis, scope for genetic testing and personalized medicine. Exp Eye Res 189:107834. doi: 10.1016/j.exer.2019.107834 [DOI] [PubMed] [Google Scholar]
- 76.Korenbrot JI (2012) Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: Facts and models. Progress in Retinal Eye Research 31:442–466. doi: 10.1016/j.preteyeres.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME (2006) RGS expression rate-limits recovery of rod photoresponses. Neuron 51:409–416. doi: 10.1016/j.neuron.2006.07.010 [DOI] [PubMed] [Google Scholar]
- 78.Lagman D, Franzen IE, Eggert J, Larhammar D, Abalo XM (2016) Evolution and expression of the phosphodiesterase 6 genes unveils vertebrate novelty to control photosensitivity. BMC Evol Biol 16:124. doi: 10.1186/s12862-016-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamb TD, Heck M, Kraft TW (2018) Implications of dimeric activation of PDE6 for rod phototransduction. Open Biol 8:180076. doi: 10.1098/rsob.180076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamb TD, Hunt DM (2017) Evolution of the vertebrate phototransduction cascade activation steps. Dev Biol 431:77–92. doi: 10.1016/j.ydbio.2017.03.018 [DOI] [PubMed] [Google Scholar]
- 81.Lamb TD, Kraft TW (2020) A quantitative account of mammalian rod phototransduction with PDE6 dimeric activation: responses to bright flashes. Open Biol 10:190241. doi: 10.1098/rsob.190241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379:311–319. doi: 10.1038/379311a0 [DOI] [PubMed] [Google Scholar]
- 83.Laties AM (2009) Vision disorders and phosphodiesterase type 5 inhibitors: a review of the evidence to date. Drug Saf 32:1–18. doi: 10.2165/00002018-200932010-00001 [DOI] [PubMed] [Google Scholar]
- 84.Leskov IB, Klenchin VA, Handy JW, Whitlock GG, Govardovskii VI, Bownds MD, Lamb TD, Pugh EN, Arshavsky VY (2000) The gain of rod phototransduction: Reconciliation of biochemical and electrophysiological measurements. Neuron 27:525–537. doi: 10.1016/s0896-6273(00)00063-5 [DOI] [PubMed] [Google Scholar]
- 85.Lishko PV, Martemyanov KA, Hopp JA, Arshavsky VY (2002) Specific binding of RGS9-Gβ5L to protein anchor in photoreceptor membranes greatly enhances its catalytic activity. J Biol Chem 277:24376–24381. doi: 10.1074/jbc.M203237200 [DOI] [PubMed] [Google Scholar]
- 86.Liu YT, Matte SL, Corbin JD, Francis SH, Cote RH (2009) Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors. Journal of Biological Chemistry 284:31541–31547. doi: 10.1074/jbc.M109.018606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Majumder A, Pahlberg J, Muradov H, Boyd KK, Sampath AP, Artemyev NO (2015) Exchange of cone for rod phosphodiesterase 6 catalytic subunits in rod photoreceptors mimics in part features of light adaptation. Journal of Neuroscience 35:9225–9235. doi: 10.1523/JNEUROSCI.3563-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martemyanov KA, Lishko PV, Calero N, Keresztes G, Sokolov M, Strissel KJ, Leskov IB, Hopp JA, Kolesnikov AV, Chen CK, Lem J, Heller S, Burns ME, Arshavsky VY (2003) The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. Journal of Neuroscience 23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez SE, Heikaus CC, Klevit RE, Beavo JA (2008) The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem 283:25913–25919. doi: 10.1074/jbc.M802891200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC (2014) Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov 13:290–314. doi: 10.1038/nrd4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melia TJ, Malinski JA, He F, Wensel TG (2000) Enhancement of phototransduction protein interactions by lipid surfaces. J Biol Chem 275:3535–3542. doi: 10.1074/jbc.275.5.3535 [DOI] [PubMed] [Google Scholar]
- 92.Michaelides M, Li Z, Rana NA, Richardson EC, Hykin PG, Moore AT, Holder GE, Webster AR (2010) Novel mutations and electrophysiologic findings in RGS9- and R9AP-associated retinal dysfunction (Bradyopsia). Ophthalmology 117:120–127 e121. doi: 10.1016/j.ophtha.2009.06.011 [DOI] [PubMed] [Google Scholar]
- 93.Min KC, Gravina SA, Sakmar TP (2000) Reconstitution of the vertebrate visual cascade using recombinant heterotrimeric transducin purified from Sf9 cells. Protein Expr Purif 20:514–526. doi: 10.1006/prep.2000.1326 [DOI] [PubMed] [Google Scholar]
- 94.Molday RS, Moritz OL (2015) Photoreceptors at a glance. J Cell Sci 128:4039–4045. doi: 10.1242/jcs.175687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mou H, Cote RH (2001) The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory γ subunit. J Biol Chem 276:27527–27534. doi: 10.1074/jbc.M103316200 [DOI] [PubMed] [Google Scholar]
- 96.Mou H, Grazio HJ, Cook TA, Beavo JA, Cote RH (1999) cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its γ and δ subunits. J Biol Chem 274:18813–18820. doi: 10.1074/jbc.274.26.18813 [DOI] [PubMed] [Google Scholar]
- 97.Muradov H, Boyd KK, Artemyev NO (2004) Structural determinants of the PDE6 GAF A domain for binding the inhibitory gamma-subunit and noncatalytic cGMP. Vision Res 44:2437–2444. doi: 10.1016/j.visres.2004.05.013 [DOI] [PubMed] [Google Scholar]
- 98.Muradov H, Boyd KK, Artemyev NO (2010) Rod phosphodiesterase-6 PDE6A and PDE6B subunits are enzymatically equivalent. J Biol Chem 285:39828–39834. doi: 10.1074/jbc.M110.170068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muradov H, Boyd KK, Haeri M, Kerov V, Knox BE, Artemyev NO (2009) Characterization of human cone phosphodiesterase-6 ectopically expressed in Xenopus laevis rods. J Biol Chem 284:32662–32669. doi: 10.1074/jbc.M109.049916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Naeem MA, Chavali VR, Ali S, Iqbal M, Riazuddin S, Khan SN, Husnain T, Sieving PA, Ayyagari R, Riazuddin S, Hejtmancik JF, Riazuddin SA (2012) GNAT1 associated with autosomal recessive congenital stationary night blindness. Invest Ophthalmol Vis Sci 53:1353–1361. doi: 10.1167/iovs.11-8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Natochin M, Granovsky AE, Artemyev NO (1998) Identification of effector residues on photoreceptor G protein, transducin. J Biol Chem 273:21808–21815. doi: 10.1074/jbc.273.34.21808 [DOI] [PubMed] [Google Scholar]
- 102.Nikolova S, Guenther A, Savai R, Weissmann N, Ghofrani HA, Konigshoff M, Eickelberg O, Klepetko W, Voswinckel R, Seeger W, Grimminger F, Schermuly RT, Pullamsetti SS (2010) Phosphodiesterase 6 subunits are expressed and altered in idiopathic pulmonary fibrosis. Respir Res 11:146. doi: 10.1186/1465-9921-11-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Norton AW, D’Amours MR, Grazio HJ, Hebert TL, Cote RH (2000) Mechanism of transducin activation of frog rod photoreceptor phosphodiesterase: allosteric interactions between the inhibitory γ subunit and the noncatalytic cGMP binding sites. Journal of Biological Chemistry 275:38611–38619. doi: 10.1074/jbc.M004606200 [DOI] [PubMed] [Google Scholar]
- 104.Paglia MJ, Mou H, Cote RH (2002) Regulation of photoreceptor phosphodiesterase (PDE6) by phosphorylation of its inhibitory γ subunit re-evaluated. Journal of Biological Chemistry 277:5017–5023. doi: 10.1074/jbc.M106328200 [DOI] [PubMed] [Google Scholar]
- 105.Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci U S A 106:18225–18230. doi: 10.1073/pnas.0907635106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pattis JG, Kamal S, Li B, May ER (2019) Catalytic Domains of Phosphodiesterase 5, 6, and 5/6 Chimera Display Differential Dynamics and Ligand Dissociation Energy Barriers. J Phys Chem B 123:825–835. doi: 10.1021/acs.jpcb.8b11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peinado Allina G, Fortenbach C, Naarendorp F, Gross OP, Pugh EN, Burns ME (2017) Bright flash response recovery of mammalian rods in vivo is rate limited by RGS9. The Journal of General Physiology 149:443–454. doi: 10.1085/jgp.201611692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pentia DC, Hosier S, Cote RH (2006) The glutamic acid-rich protein-2 (GARP2) is a high affinity rod photoreceptor phosphodiesterase (PDE6)-binding protein that modulates its catalytic properties. Journal of Biological Chemistry 281:5500–5505. doi: 10.1074/jbc.M507488200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pittler SJ, Baehr W (1991) Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase β-subunit gene of the rd mouse. Proc Natl Acad Sci U S A 88:8322–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ponzoni L, Peñaherrera DA, Oltvai ZN, Bahar I (2020) Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics 36:3084–3092. doi: 10.1093/bioinformatics/btaa127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Power M, Das S, Schutze K, Marigo V, Ekstrom P, Paquet-Durand F (2020) Cellular mechanisms of hereditary photoreceptor degeneration - Focus on cGMP. Prog Retin Eye Res 74:100772. doi: 10.1016/j.preteyeres.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 112.Pugh EN, Lamb TD (1993) Amplification and kinetics of the activation steps in phototransduction. Biochim Biophys Acta 1141:111–149. doi: 10.1016/0005-2728(93)90038-h [DOI] [PubMed] [Google Scholar]
- 113.Qi C, Sorrentino S, Medalia O, Korkhov VM (2019) The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein. Science 364:389–394. doi: 10.1126/science.aav0778 [DOI] [PubMed] [Google Scholar]
- 114.Qureshi BM, Behrmann E, Schoneberg J, Loerke J, Burger J, Mielke T, Giesebrecht J, Noe F, Lamb TD, Hofmann KP, Spahn CMT, Heck M (2018) It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods. Open Biol 8:180075 180010.181098/rsob.180075. doi: 10.1098/rsob.180075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reingruber J, Ingram NT, Griffis KG, Fain GL (2020) A kinetic analysis of mouse rod and cone photoreceptor responses. J Physiol 598:3747–3763. doi: 10.1113/JP279524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Remmer MH, Rastogi N, Ranka MP, Ceisler EJ (2015) Achromatopsia: a review. Curr Opin Ophthalmol 26:333–340. doi: 10.1097/ICU.0000000000000189 [DOI] [PubMed] [Google Scholar]
- 117.Rieke F, Baylor DA (1996) Molecular origin of continuous dark noise in rod photoreceptors. Biophys J 71:2553–2572. doi: 10.1016/S0006-3495(96)79448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rieke F, Baylor DA (2000) Origin and functional impact of dark noise in retinal cones. Neuron 26:181–186. doi: 10.1016/s0896-6273(00)81148-4 [DOI] [PubMed] [Google Scholar]
- 119.Sidman RL, Green MC (1965) Retinal degeneration in the mouse: Location of the rd Locus in Linkage Group XVII. Journal of Heredity 56:23–29. doi: 10.1093/oxfordjournals.jhered.a107364 [DOI] [PubMed] [Google Scholar]
- 120.Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL (2008) Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci U S A 105:1505–1510. doi: 10.1073/pnas.0709558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Strauss RW, Dubis AM, Cooper RF, Ba-Abbad R, Moore AT, Webster AR, Dubra A, Carroll J, Michaelides M (2015) Retinal Architecture in RGS9- and R9AP-Associated Retinal Dysfunction (Bradyopsia). Am J Ophthalmol 160:1269–1275 e1261. doi: 10.1016/j.ajo.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Szabo V, Kreienkamp HJ, Rosenberg T, Gal A (2007) p.Gln200Glu, a putative constitutively active mutant of rod a-transducin (GNAT1) in autosomal dominant congenital stationary night blindness. Hum Mutat 28:741–742. doi: 10.1002/humu.9499 [DOI] [PubMed] [Google Scholar]
- 123.Tate RJ, Arshavsky VY, Pyne NJ (2002) The identification of the inhibitory gamma-subunits of the type 6 retinal cyclic guanosine monophosphate phosphodiesterase in non-retinal tissues: differential processing of mRNA transcripts. Genomics 79:582–586. doi: 10.1006/geno.2002.6740 [DOI] [PubMed] [Google Scholar]
- 124.Thompson DA, Iannaccone A, Ali RR, Arshavsky VY, Audo I, Bainbridge JWB, Besirli CG, Birch DG, Branham KE, Cideciyan AV, Daiger SP, Dalkara D, Duncan JL, Fahim AT, Flannery JG, Gattegna R, Heckenlively JR, Heon E, Jayasundera KT, Khan NW, Klassen H, Leroy BP, Molday RS, Musch DC, Pennesi ME, Petersen-Jones SM, Pierce EA, Rao RC, Reh TA, Sahel JA, Sharon D, Sieving PA, Strettoi E, Yang P, Zacks DN, Monaciano C (2020) Advancing Clinical Trials for Inherited Retinal Diseases: Recommendations from the Second Monaciano Symposium. Transl Vis Sci Technol 9:2. doi: 10.1167/tvst.9.7.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tian M, Zallocchi M, Wang W, Chen CK, Palczewski K, Delimont D, Cosgrove D, Peng YW (2013) Light-induced translocation of RGS9–1 and Gβ5L in mouse rod photoreceptors. PLoS One 8:e58832. doi: 10.1371/journal.pone.0058832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tolone A, Belhadj S, Rentsch A, Schwede F, Paquet-Durand F (2019) The cGMP Pathway and Inherited Photoreceptor Degeneration: Targets, Compounds, and Biomarkers. Genes (Basel) 10:453. doi: 10.3390/genes10060453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsang SH, Sharma T (2018) Congenital Stationary Night Blindness. Adv Exp Med Biol 1085:61–64. doi: 10.1007/978-3-319-95046-4_13 [DOI] [PubMed] [Google Scholar]
- 128.Tsang SH, Sharma T (2018) Progressive Cone Dystrophy and Cone-Rod Dystrophy (XL, AD, and AR). Adv Exp Med Biol 1085:53–60. doi: 10.1007/978-3-319-95046-4_12 [DOI] [PubMed] [Google Scholar]
- 129.Tsang SH, Woodruff ML, Janisch KM, Cilluffo MC, Farber DB, Fain GL (2007) Removal of phosphorylation sites of γ subunit of phosphodiesterase 6 alters rod light response. Journal of Physiology 579:303–312. doi: 10.1113/jphysiol.2006.121772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A (2015) Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech 8:109–129. doi: 10.1242/dmm.017913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang H, Robinson H, Ke H (2010) Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J Biol Chem 285:38149–38156. doi: 10.1074/jbc.M110.141614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang T, Tsang SH, Chen J (2017) Two pathways of rod photoreceptor cell death induced by elevated cGMP. Hum Mol Genet 26:2299–2306. doi: 10.1093/hmg/ddx121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, Plachetzki DC, Cote RH (2019) The N termini of the inhibitory γ-subunits of phosphodiesterase-6 (PDE6) from rod and cone photoreceptors differentially regulate transducin-mediated PDE6 activation. J Biol Chem 294:8351–8360. doi: 10.1074/jbc.RA119.007520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Webb B, Viswanath S, Bonomi M, Pellarin R, Greenberg CH, Saltzberg D, Sali A (2018) Integrative structure modeling with the Integrative Modeling Platform. Protein Sci 27:245–258. doi: 10.1002/pro.3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wen XH, Dizhoor AM, Makino CL (2014) Membrane guanylyl cyclase complexes shape the photoresponses of retinal rods and cones. Front Mol Neurosci 7:45. doi: 10.3389/fnmol.2014.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wensel TG (2008) Signal transducing membrane complexes of photoreceptor outer segments. Vision Res 48:2052–2061. doi: 10.1016/j.visres.2008.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wensel TG, Stryer L (1990) Activation mechanism of retinal rod cyclic GMP phosphodiesterase probed by fluorescein-labeled inhibitory subunit. Biochemistry 29:2155–2161. doi: 10.1021/bi00460a028 [DOI] [PubMed] [Google Scholar]
- 138.Woodruff ML, Janisch KM, Peshenko IV, Dizhoor AM, Tsang SH, Fain GL (2008) Modulation of phosphodiesterase6 turnoff during background illumination in mouse rod photoreceptors. Journal of Neuroscience 28:2064–2074. doi: 10.1523/JNEUROSCI.2973-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yadav RP, Artemyev NO (2017) AIPL1: A specialized chaperone for the phototransduction effector. Cell Signal 40:183–189. doi: 10.1016/j.cellsig.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamazaki A, Bartucci F, Ting A, Bitensky MW (1982) Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc Natl Acad Sci U S A 79:3702–3706. doi: 10.1073/pnas.79.12.3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yamazaki A, Bondarenko VA, Dua S, Yamazaki M, Usukura J, Hayashi F (1996) Possible stimulation of retinal rod recovery to dark state by cGMP release from a cGMP phosphodiesterase noncatalytic site. J Biol Chem 271:32495–32498. doi: 10.1074/jbc.271.51.32495 [DOI] [PubMed] [Google Scholar]
- 142.Yue WWS, Silverman D, Ren X, Frederiksen R, Sakai K, Yamashita T, Shichida Y, Cornwall MC, Chen J, Yau KW (2019) Elementary response triggered by transducin in retinal rods. Proc Natl Acad Sci U S A 116:5144–5153. doi: 10.1073/pnas.1817781116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zeng-Elmore X, Gao XZ, Pellarin R, Schneidman-Duhovny D, Zhang XJ, Kozacka KA, Tang Y, Sali A, Chalkley RJ, Cote RH, Chu F (2014) Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J Mol Biol 426:3713–3728. doi: 10.1016/j.jmb.2014.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang XJ, Cahill KB, Elfenbein A, Arshavsky VY, Cote RH (2008) Direct Allosteric Regulation between the GAF Domain and Catalytic Domain of Photoreceptor Phosphodiesterase PDE6. Journal of Biological Chemistry 283:29699–29705. doi: 10.1074/jbc.M803948200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang XJ, Feng Q, Cote RH (2005) Efficacy and selectivity of phosphodiesterase-targeted drugs in inhibiting photoreceptor phosphodiesterase (PDE6) in retinal photoreceptors. Investigative Ophthalmology & Visual Science 46:3060–3066. doi: 10.1167/iovs.05-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang XJ, Gao XZ, Yao W, Cote RH (2012) Functional mapping of interacting regions of the photoreceptor phosphodiesterase (PDE6) γ-subunit with PDE6 catalytic dimer, transducin, and Regulator of G-protein Signaling9–1 (RGS9–1). J Biol Chem 287:26312–26320. doi: 10.1074/jbc.M112.377333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang XJ, Skiba NP, Cote RH (2010) Structural requirements of the photoreceptor phosphodiesterase gamma-subunit for inhibition of rod PDE6 holoenzyme and for its activation by transducin. J Biol Chem 285:4455–4463. doi: 10.1074/jbc.M109.057406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang Z, He F, Constantine R, Baker ML, Baehr W, Schmid MF, Wensel TG, Agosto MA (2015) Domain organization and conformational plasticity of the G protein effector, PDE6. J Biol Chem 290:12833–12843. doi: 10.1074/jbc.A115.647636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zoraghi R, Corbin JD, Francis SH (2004) Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol Pharmacol 65:267–278. doi: 10.1124/mol.65.2.267 [DOI] [PubMed] [Google Scholar]
- 150.Zuo H, Faiz A, van den Berge M, Mudiyanselage S, Borghuis T, Timens W, Nikolaev VO, Burgess JK, Schmidt M (2020) Cigarette smoke exposure alters phosphodiesterases in human structural lung cells. Am J Physiol Lung Cell Mol Physiol 318:L59–L64. doi: 10.1152/ajplung.00319.2019 [DOI] [PubMed] [Google Scholar]