Figure 4. DA sensing stability with 3DFG and CFEs.
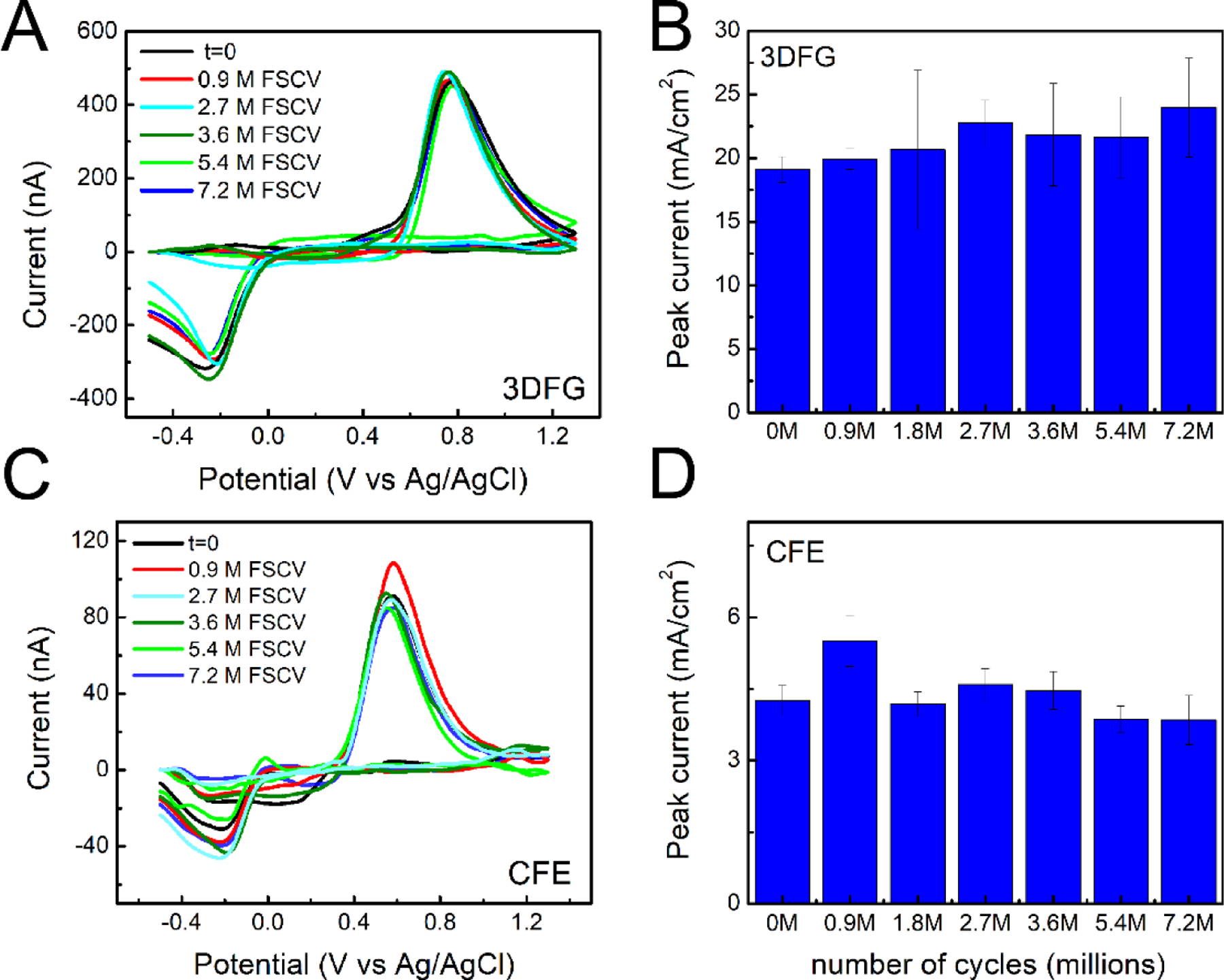
(A,B) 3DFG: (A) Representative background subtracted CVs in response to 1μM bolus injections of DA at the time 0 and after 25, 50, 75,100, 150 and 200 hours of 3DFG FSCV continuous scanning in PBS. (B) Column plot reporting the amplitude of the oxidation peaks collected in response to 1μM bolus injections of DA at time zero and after 0.9, 2.7, 3.6, 5.4 and 7.2 million FSCV cycles (corresponding to 25, 50, 75,100, 150 and 200 h) of 3DFG FSCV continuous scanning in PBS. (Repeated measurements ANOVA, F(6,30)=2.07899, P= 0.08559>0.05, Bonferroni post-hoc test ns) (C, D) CFEs: (C) Representative background subtracted CVs in response to 1μM bolus injections of DA at the time 0 and after 0.9, 2.7, 3.6, 5.4 and 7.2 million FSCV cycles of CFE FSCV continuous scanning in PBS. (D) Column plot reporting the amplitude of the oxidation peaks collected in response to 1μM bolus injections of DA at time zero and after 0.9, 2.7, 3.6, 5.4 and 7.2 million FSCV cycles (i.e. 25, 50, 75,100, 150 and 200 h) of CFE FSCV continuous scanning in PBS. (Repeated measurements with ANOVA, F(6,36)=2.04792, P=0.08432>0.05, Bonferroni post-test ns).
